+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Sub-tomogram average of the C. elegans ATP synthase dimer | |||||||||
 Map data Map data | Post-processed sub-tomogram averaging map of the C. elegans ATP synthase dimer at a nominal resolution of 38.6 Angstrom. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Supercomplex / dimer / Supercomplex / dimer /  enzyme / respiration. / enzyme / respiration. /  MEMBRANE PROTEIN MEMBRANE PROTEIN | |||||||||
| Biological species |   Caenorhabditis elegans (invertebrata) Caenorhabditis elegans (invertebrata) | |||||||||
| Method | subtomogram averaging /  cryo EM / Resolution: 38.6 Å cryo EM / Resolution: 38.6 Å | |||||||||
 Authors Authors | Buzzard EJ / McLaren M / Gold VAM | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Biochem J / Year: 2024 Journal: Biochem J / Year: 2024Title: The consequence of ATP synthase dimer angle on mitochondrial morphology studied by cryo-electron tomography. Authors: Emma Buzzard / Mathew McLaren / Piotr Bragoszewski / Andrea Brancaccio / Holly Ford / Bertram Daum / Patricia Kuwabara / Ian Collinson / Vicki Gold /    Abstract: Mitochondrial ATP synthases form rows of dimers, which induce membrane curvature to give cristae their characteristic lamellar or tubular morphology. The angle formed between the central stalks of ...Mitochondrial ATP synthases form rows of dimers, which induce membrane curvature to give cristae their characteristic lamellar or tubular morphology. The angle formed between the central stalks of ATP synthase dimers varies between species. Using cryo-electron tomography and sub-tomogram averaging, we determined the structure of the ATP synthase dimer from the nematode worm C. elegans and show that the dimer angle differs from previously determined structures. The consequences of this species-specific difference at the dimer interface were investigated by comparing C. elegans and S. cerevisiae mitochondrial morphology. We reveal that C. elegans has a larger ATP synthase dimer angle with more lamellar (flatter) cristae when compared to yeast. The underlying cause of this difference was investigated by generating an atomic model of the C. elegans ATP synthase dimer by homology modelling. A comparison of our C. elegans model to an existing S. cerevisiae structure reveals the presence of extensions and rearrangements in C. elegans subunits associated with maintaining the dimer interface. We speculate that increasing dimer angles could provide an advantage for species that inhabit variable-oxygen environments by forming flatter more energetically efficient cristae. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18991.map.gz emd_18991.map.gz | 2.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18991-v30.xml emd-18991-v30.xml emd-18991.xml emd-18991.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18991_fsc.xml emd_18991_fsc.xml | 5.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_18991.png emd_18991.png | 53.9 KB | ||
| Masks |  emd_18991_msk_1.map emd_18991_msk_1.map | 10.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18991.cif.gz emd-18991.cif.gz | 4.7 KB | ||
| Others |  emd_18991_half_map_1.map.gz emd_18991_half_map_1.map.gz emd_18991_half_map_2.map.gz emd_18991_half_map_2.map.gz | 7.3 MB 7.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18991 http://ftp.pdbj.org/pub/emdb/structures/EMD-18991 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18991 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18991 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18991.map.gz / Format: CCP4 / Size: 10.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18991.map.gz / Format: CCP4 / Size: 10.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post-processed sub-tomogram averaging map of the C. elegans ATP synthase dimer at a nominal resolution of 38.6 Angstrom. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.58 Å | ||||||||||||||||||||||||||||||||||||
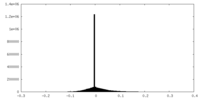
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18991_msk_1.map emd_18991_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
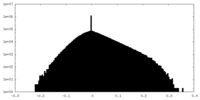
| Density Histograms |
-Half map: Half map 1 from relion refinement job used...
| File | emd_18991_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 from relion refinement job used to generate final post-processed map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2 from relion refinement job used...
| File | emd_18991_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 from relion refinement job used to generate final post-processed map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : C. elegans mitochondrial ATP synthase dimer
| Entire | Name: C. elegans mitochondrial ATP synthase dimer |
|---|---|
| Components |
|
-Supramolecule #1: C. elegans mitochondrial ATP synthase dimer
| Supramolecule | Name: C. elegans mitochondrial ATP synthase dimer / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Caenorhabditis elegans (invertebrata) / Strain: N2 (Bristol) / Organ: Whole organism / Tissue: Whole organism / Organelle: Mitochondria / Location in cell: Inner mitochondrial membrane Caenorhabditis elegans (invertebrata) / Strain: N2 (Bristol) / Organ: Whole organism / Tissue: Whole organism / Organelle: Mitochondria / Location in cell: Inner mitochondrial membrane |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: UltrAuFoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV | |||||||||||||||
| Details | Mitochondria isolated from C. elegans underwent 2-3 freeze-thaw cycles, to break open the mitochondria and release the cristae. Tomography was completed using these crista membranes. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 7.0 µm / Nominal defocus min: 5.0 µm / Nominal magnification: 39000 Bright-field microscopy / Nominal defocus max: 7.0 µm / Nominal defocus min: 5.0 µm / Nominal magnification: 39000 |
| Specialist optics | Energy filter - Name: GIF Quantum LS |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Number real images: 3 / Average exposure time: 2.8 sec. / Average electron dose: 1.36 e/Å2 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)