+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
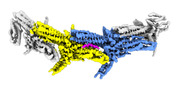
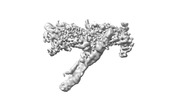
| Title | Ndc80c microtubule complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Kinetochore / Kinetochore /  microtubule / error correction / microtubule / error correction /  chromosome segregation / chromosome segregation /  CELL CYCLE CELL CYCLE | |||||||||
| Biological species |   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.11 Å cryo EM / Resolution: 4.11 Å | |||||||||
 Authors Authors | Muir KW / Barford D | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2023 Journal: Science / Year: 2023Title: Structural mechanism of outer kinetochore Dam1-Ndc80 complex assembly on microtubules. Authors: Kyle W Muir / Christopher Batters / Tom Dendooven / Jing Yang / Ziguo Zhang / Alister Burt / David Barford /  Abstract: Kinetochores couple chromosomes to the mitotic spindle to segregate the genome during cell division. An error correction mechanism drives the turnover of kinetochore-microtubule attachments until ...Kinetochores couple chromosomes to the mitotic spindle to segregate the genome during cell division. An error correction mechanism drives the turnover of kinetochore-microtubule attachments until biorientation is achieved. The structural basis for how kinetochore-mediated chromosome segregation is accomplished and regulated remains an outstanding question. In this work, we describe the cryo-electron microscopy structure of the budding yeast outer kinetochore Ndc80 and Dam1 ring complexes assembled onto microtubules. Complex assembly occurs through multiple interfaces, and a staple within Dam1 aids ring assembly. Perturbation of key interfaces suppresses yeast viability. Force-rupture assays indicated that this is a consequence of impaired kinetochore-microtubule attachment. The presence of error correction phosphorylation sites at Ndc80-Dam1 ring complex interfaces and the Dam1 staple explains how kinetochore-microtubule attachments are destabilized and reset. #1:  Journal: To Be Published Journal: To Be PublishedTitle: Mechanism of outer kinetochore assembly on microtubules and its regulation by mitotic error correction Authors: Muir KW / Batters C / Dendooven T / Yang J / Zhang Z / Burt A / Barford D | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18485.map.gz emd_18485.map.gz | 482.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18485-v30.xml emd-18485-v30.xml emd-18485.xml emd-18485.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_18485.png emd_18485.png | 168.9 KB | ||
| Filedesc metadata |  emd-18485.cif.gz emd-18485.cif.gz | 4.3 KB | ||
| Others |  emd_18485_half_map_1.map.gz emd_18485_half_map_1.map.gz emd_18485_half_map_2.map.gz emd_18485_half_map_2.map.gz | 482.7 MB 482.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18485 http://ftp.pdbj.org/pub/emdb/structures/EMD-18485 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18485 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18485 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18485.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18485.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_18485_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_18485_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Outer kinetochore Dam1 protomer monomer with staple and Ndc80-Nuf...
| Entire | Name: Outer kinetochore Dam1 protomer monomer with staple and Ndc80-Nuf2 coiled-coils |
|---|---|
| Components |
|
-Supramolecule #1: Outer kinetochore Dam1 protomer monomer with staple and Ndc80-Nuf...
| Supramolecule | Name: Outer kinetochore Dam1 protomer monomer with staple and Ndc80-Nuf2 coiled-coils type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#12 |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) |
| Molecular weight | Theoretical: 673772.36 MDa |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.2 µm Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.2 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD Software: (Name: cryoSPARC (ver. 3.3.2), RELION (ver. 4.0-dev)) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 4.2) |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.11 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 110704 |
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Details | Initial rigid body fitting was performed in chimera, with manual correction in coot and real-space refinement in PHENIX |
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 540.98 |
 Movie
Movie Controller
Controller






 Z
Z Y
Y X
X

















