+ Open data
Open data
- Basic information
Basic information
| Entry | 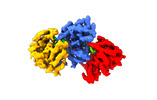 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Focused reconstruction of influenza A RNP-like particle | |||||||||
 Map data Map data | Focused reconstruction map post-processed. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Influenza virus / Influenza virus /  Nucleocapsid-like / Nucleocapsid-like /  RNA binding protein. / RNA binding protein. /  VIRAL PROTEIN VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationhelical viral capsid / viral penetration into host nucleus / viral nucleocapsid / symbiont entry into host cell /  ribonucleoprotein complex / host cell nucleus / structural molecule activity / ribonucleoprotein complex / host cell nucleus / structural molecule activity /  RNA binding / identical protein binding RNA binding / identical protein bindingSimilarity search - Function | |||||||||
| Biological species |    Influenza A virus / synthetic construct (others) Influenza A virus / synthetic construct (others) | |||||||||
| Method | helical reconstruction /  cryo EM / Resolution: 5.3 Å cryo EM / Resolution: 5.3 Å | |||||||||
 Authors Authors | Chenavier F / Estrozi LF / Zarkadas E / Ruigrok RWH / Schoehn G / Ballandras-Colas A / Crepin T | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Cryo-EM structure of influenza helical nucleocapsid reveals NP-NP and NP-RNA interactions as a model for the genome encapsidation. Authors: Florian Chenavier / Leandro F Estrozi / Jean-Marie Teulon / Eleftherios Zarkadas / Lily-Lorette Freslon / Jean-Luc Pellequer / Rob W H Ruigrok / Guy Schoehn / Allison Ballandras-Colas / Thibaut Crépin /  Abstract: Influenza virus genome encapsidation is essential for the formation of a helical viral ribonucleoprotein (vRNP) complex composed of nucleoproteins (NP), the trimeric polymerase, and the viral genome. ...Influenza virus genome encapsidation is essential for the formation of a helical viral ribonucleoprotein (vRNP) complex composed of nucleoproteins (NP), the trimeric polymerase, and the viral genome. Although low-resolution vRNP structures are available, it remains unclear how the viral RNA is encapsidated and how NPs assemble into the helical filament specific of influenza vRNPs. In this study, we established a biological tool, the RNP-like particles assembled from recombinant influenza A virus NP and synthetic RNA, and we present the first subnanometric cryo-electron microscopy structure of the helical NP-RNA complex (8.7 to 5.3 Å). The helical RNP-like structure reveals a parallel double-stranded conformation, allowing the visualization of NP-NP and NP-RNA interactions. The RNA, located at the interface of neighboring NP protomers, interacts with conserved residues previously described as essential for the NP-RNA interaction. The NP undergoes conformational changes to enable RNA binding and helix formation. Together, our findings provide relevant insights for understanding the mechanism for influenza genome encapsidation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18044.map.gz emd_18044.map.gz | 4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18044-v30.xml emd-18044-v30.xml emd-18044.xml emd-18044.xml | 23.1 KB 23.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18044_fsc.xml emd_18044_fsc.xml | 3.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_18044.png emd_18044.png | 50.7 KB | ||
| Masks |  emd_18044_msk_1.map emd_18044_msk_1.map | 4.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18044.cif.gz emd-18044.cif.gz | 7.1 KB | ||
| Others |  emd_18044_additional_1.map.gz emd_18044_additional_1.map.gz emd_18044_half_map_1.map.gz emd_18044_half_map_1.map.gz emd_18044_half_map_2.map.gz emd_18044_half_map_2.map.gz | 2.3 MB 4.2 MB 4.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18044 http://ftp.pdbj.org/pub/emdb/structures/EMD-18044 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18044 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18044 | HTTPS FTP |
-Related structure data
| Related structure data |  8pzqMC  8pzpC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18044.map.gz / Format: CCP4 / Size: 4.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18044.map.gz / Format: CCP4 / Size: 4.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused reconstruction map post-processed. | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.68 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18044_msk_1.map emd_18044_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Focused reconstruction map non post-processed.
| File | emd_18044_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused reconstruction map non post-processed. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 1.
| File | emd_18044_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map_1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 2.
| File | emd_18044_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map_2. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Influenza A NP-RNA complex
| Entire | Name: Influenza A NP-RNA complex |
|---|---|
| Components |
|
-Supramolecule #1: Influenza A NP-RNA complex
| Supramolecule | Name: Influenza A NP-RNA complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: Nucleoprotein
| Supramolecule | Name: Nucleoprotein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:    Influenza A virus Influenza A virus |
-Supramolecule #3: RNA (5'P-(UC)6-FAM3')
| Supramolecule | Name: RNA (5'P-(UC)6-FAM3') / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: Nucleoprotein
| Macromolecule | Name: Nucleoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Influenza A virus Influenza A virus |
| Molecular weight | Theoretical: 57.525965 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: MATKGTKRSY EQMETDGERQ NATEIRASVG KMIDGIGRFY IQMCTELKLS DYEGRLIQNS LTIERMVLSA FDERRNKYLE EHPSAGKDP KKTGGPIYRR VDGKWRRELI LYDKEEIRRI WRQANNGDDA TAGLTHMMIW HSNLNDATYQ RTRALVRTGM D PRMCSLMQ ...String: MATKGTKRSY EQMETDGERQ NATEIRASVG KMIDGIGRFY IQMCTELKLS DYEGRLIQNS LTIERMVLSA FDERRNKYLE EHPSAGKDP KKTGGPIYRR VDGKWRRELI LYDKEEIRRI WRQANNGDDA TAGLTHMMIW HSNLNDATYQ RTRALVRTGM D PRMCSLMQ GSTLPRRSGA AGAAVKGVGT MVMELIRMIK RGINDRNFWR GENGRRTRIA YERMCNILKG KFQTAAQRTM VD QVRESRN PGNAEFEDLI FLARSALILR GSVAHKSCLP ACVYGSAVAS GYDFEREGYS LVGIDPFRLL QNSQVYSLIR PNE NPAHKS QLVWMACHSA AFEDLRVSSF IRGTKVVPRG KLSTRGVQIA SNENMETMES STLELRSRYW AIRTRSGGNT NQQR ASSGQ ISIQPTFSVQ RNLPFDRPTI MAAFTGNTEG RTSDMRTEII RLMESARPED VSFQGRGVFE LSDEKATSPI VPSFD MSNE GSYFFGDNAE EYDNLEHHHH HH UniProtKB:  Nucleoprotein Nucleoprotein |
-Macromolecule #2: RNA (5'P-(UC)6-FAM3')
| Macromolecule | Name: RNA (5'P-(UC)6-FAM3') / type: rna / ID: 2 / Number of copies: 2 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 3.623128 KDa |
| Sequence | String: UCUCUCUCUC UC |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.23 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: 20 mM HEPES 150 mM NaCl 5 mM beta-mercaptoethanol 2 mM methyl-PEG8-NHS | ||||||||||||||||||
| Grid | Model: Quantifoil R0.6/1 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Details: 25 mA | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||||||||
| Details | 20 mM HEPES 150 mM NaCl 5 mM beta-mercaptoethanol Incubation overnight with 0.004 uM RNA (5'P-(UC)6-FAM3') Prior freezing, the sample was incubated 30 min on ice with 2 mM methyl-PEG8-NHS |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 2.2 µm / Calibrated defocus min: 0.8 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Specialist optics | Energy filter - Name: GIF Quantum LS |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 26515 / Average exposure time: 1.9 sec. / Average electron dose: 40.8 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Residue range: 21-490 / Chain - Source name: PDB / Chain - Initial model type: experimental model / Details: Fitting of the NPcore by using ChimeraX |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-8pzq: |
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X



































