+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Complex of HK97 small terminase with DNA | |||||||||
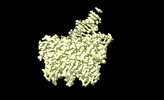
 Map data Map data | HK97 small terminase in complex with DNA | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HK97 small terminase /  bacteriophage / protein:DNA complex / cryoEM structure / bacteriophage / protein:DNA complex / cryoEM structure /  DNA BINDING PROTEIN DNA BINDING PROTEIN | |||||||||
| Function / homology | Terminase small subunit Function and homology information Function and homology information | |||||||||
| Biological species |   Byrnievirus HK97 Byrnievirus HK97 | |||||||||
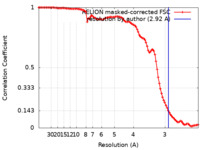
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.92 Å cryo EM / Resolution: 2.92 Å | |||||||||
 Authors Authors | Chechik M / Greive SJ / Antson AA / Jenkins HT | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2023 Journal: bioRxiv / Year: 2023Title: Structure of HK97 small terminase:DNA complex unveils a novel DNA binding mechanism by a circular protein. Authors: Maria Chechik / Sandra J Greive / Alfred A Antson / Huw T Jenkins /  Abstract: DNA recognition is critical for assembly of double-stranded DNA viruses, in particular for the initiation of packaging the viral genome into the capsid. DNA packaging has been extensively studied for ...DNA recognition is critical for assembly of double-stranded DNA viruses, in particular for the initiation of packaging the viral genome into the capsid. DNA packaging has been extensively studied for three archetypal bacteriophage systems: , and phi29. We identified the minimal site within the region of bacteriophage HK97 specifically recognised by the small terminase and determined a cryoEM structure for the small terminase:DNA complex. This nonameric circular protein utilizes a previously unknown mechanism of DNA binding. While DNA threads through the central tunnel, unexpectedly, DNA-recognition is generated at its exit by a substructure formed by the N- and C-terminal segments of two adjacent protomers of the terminase which are unstructured in the absence of DNA. Such interaction ensures continuous engagement of the small terminase with DNA, allowing sliding along DNA while simultaneously checking the DNA sequence. This mechanism allows locating and instigating packaging initiation and termination precisely at the site. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17818.map.gz emd_17818.map.gz | 7.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17818-v30.xml emd-17818-v30.xml emd-17818.xml emd-17818.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17818_fsc.xml emd_17818_fsc.xml | 10.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_17818.png emd_17818.png | 54.2 KB | ||
| Masks |  emd_17818_msk_1.map emd_17818_msk_1.map | 8 MB |  Mask map Mask map | |
| Others |  emd_17818_half_map_1.map.gz emd_17818_half_map_1.map.gz emd_17818_half_map_2.map.gz emd_17818_half_map_2.map.gz | 7.4 MB 7.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17818 http://ftp.pdbj.org/pub/emdb/structures/EMD-17818 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17818 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17818 | HTTPS FTP |
-Related structure data
| Related structure data |  8popC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17818.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17818.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HK97 small terminase in complex with DNA | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.18889 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17818_msk_1.map emd_17818_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: HK97 small terminase in complex with DNA
| File | emd_17818_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HK97 small terminase in complex with DNA | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: HK97 small terminase in complex with DNA
| File | emd_17818_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HK97 small terminase in complex with DNA | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : complex of HK97 with DNA
| Entire | Name: complex of HK97 with DNA |
|---|---|
| Components |
|
-Supramolecule #1: complex of HK97 with DNA
| Supramolecule | Name: complex of HK97 with DNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Byrnievirus HK97 Byrnievirus HK97 |
| Molecular weight | Theoretical: 185 KDa |
-Macromolecule #1: small terminase
| Macromolecule | Name: small terminase / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Byrnievirus HK97 Byrnievirus HK97 |
| Recombinant expression | Organism:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) |
| Sequence | String: MADKRIRSDS SAAAVQAMKN AAVDTIDPPS HAGLEKKAEP FWHDNIRSKA LDSWTPADLL AAVELANNQ LYITVLRKDL RKEERIRGEE RDEGLIKDLR KQIVELQRTI LAQRRDLQIH S HATNGESR DQKKRNQNDR DARNTKNEHQ DQDDNLIAFP KHG UniProtKB: Terminase small subunit |
-Macromolecule #2: DNA (31-mer)
| Macromolecule | Name: DNA (31-mer) / type: dna / ID: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Byrnievirus HK97 Byrnievirus HK97 |
| Sequence | String: TAAAACTAAA AAAATCGGGT TAGCGTTAAA T |
-Macromolecule #3: DNA (31-mer)
| Macromolecule | Name: DNA (31-mer) / type: dna / ID: 3 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Byrnievirus HK97 Byrnievirus HK97 |
| Sequence | String: ATTTAACGCT AACCCGATTT TTTTAGTTTT A |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 180 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.038 kPa | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | complex was purified on S200 10/300 column |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 2767 / Average exposure time: 8.0 sec. / Average electron dose: 53.6 e/Å2 Details: Two datasets were collected: 1st dataset: 682 images 2nd dataset: 2085 images |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X