[English] 日本語
 Yorodumi
Yorodumi- EMDB-17756: Structure of the murine trace amine-associated receptor TAAR7f bo... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the murine trace amine-associated receptor TAAR7f bound to N,N-dimethylcyclohexylamine (DMCH) in complex with mini-Gs trimeric G protein | |||||||||
 Map data Map data | Consensus map of the mTAAR7f receptor bound to NDMCH in complex with miniGs trimer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | trace-amine associated receptor / TAAR / mTAAR7f /  GPCR / GPCR /  receptor / receptor /  G protein / G protein /  MEMBRANE PROTEIN MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationtrace-amine receptor activity / sensory perception of chemical stimulus / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels ...trace-amine receptor activity / sensory perception of chemical stimulus / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Adrenaline,noradrenaline inhibits insulin secretion / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / cellular response to catecholamine stimulus / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / sensory perception of taste / GPER1 signaling / G-protein beta-subunit binding /  heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade /  extracellular vesicle / G alpha (12/13) signalling events / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye / extracellular vesicle / G alpha (12/13) signalling events / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye /  GTPase binding / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (i) signalling events / fibroblast proliferation / G alpha (s) signalling events / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase binding / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (i) signalling events / fibroblast proliferation / G alpha (s) signalling events / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling / G protein-coupled receptor signaling pathway / lysosomal membrane /  GTPase activity / GTPase activity /  synapse / protein-containing complex binding / synapse / protein-containing complex binding /  signal transduction / extracellular exosome / signal transduction / extracellular exosome /  membrane / membrane /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /   Lama glama (llama) / Lama glama (llama) /   Mus musculus (house mouse) Mus musculus (house mouse) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.92 Å cryo EM / Resolution: 2.92 Å | |||||||||
 Authors Authors | Gusach A / Lee Y / Edwards PC / Huang F / Weyand SN / Tate CG | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2023 Journal: bioRxiv / Year: 2023Title: Molecular recognition of an aversive odorant by the murine trace amine-associated receptor TAAR7f. Authors: Anastasiia Gusach / Yang Lee / Armin Nikpour Khoshgrudi / Elizaveta Mukhaleva / Ning Ma / Eline J Koers / Qingchao Chen / Patricia C Edwards / Fanglu Huang / Jonathan Kim / Filippo Mancia / ...Authors: Anastasiia Gusach / Yang Lee / Armin Nikpour Khoshgrudi / Elizaveta Mukhaleva / Ning Ma / Eline J Koers / Qingchao Chen / Patricia C Edwards / Fanglu Huang / Jonathan Kim / Filippo Mancia / Dmitry B Verprintsev / Nagarajan Vaidehi / Simone N Weyand / Christopher G Tate /   Abstract: There are two main families of G protein-coupled receptors that detect odours in humans, the odorant receptors (ORs) and the trace amine-associated receptors (TAARs). Their amino acid sequences are ...There are two main families of G protein-coupled receptors that detect odours in humans, the odorant receptors (ORs) and the trace amine-associated receptors (TAARs). Their amino acid sequences are distinct, with the TAARs being most similar to the aminergic receptors such as those activated by adrenaline, serotonin and histamine. To elucidate the structural determinants of ligand recognition by TAARs, we have determined the cryo-EM structure of a murine receptor, mTAAR7f, coupled to the heterotrimeric G protein G and bound to the odorant N,N-dimethylcyclohexylamine (DMCH) to an overall resolution of 2.9 Å. DMCH is bound in a hydrophobic orthosteric binding site primarily through van der Waals interactions and a strong charge-charge interaction between the tertiary amine of the ligand and an aspartic acid residue. This site is distinct and non-overlapping with the binding site for the odorant propionate in the odorant receptor OR51E2. The structure, in combination with mutagenesis data and molecular dynamics simulations suggests that the activation of the receptor follows a similar pathway to that of the β-adrenoceptors, with the significant difference that DMCH interacts directly with one of the main activation microswitch residues. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17756.map.gz emd_17756.map.gz | 93 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17756-v30.xml emd-17756-v30.xml emd-17756.xml emd-17756.xml | 28.9 KB 28.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17756_fsc.xml emd_17756_fsc.xml | 12.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_17756.png emd_17756.png | 121 KB | ||
| Masks |  emd_17756_msk_1.map emd_17756_msk_1.map emd_17756_msk_2.map emd_17756_msk_2.map | 196.4 MB 196.4 MB |  Mask map Mask map | |
| Others |  emd_17756_additional_1.map.gz emd_17756_additional_1.map.gz emd_17756_half_map_1.map.gz emd_17756_half_map_1.map.gz emd_17756_half_map_2.map.gz emd_17756_half_map_2.map.gz | 93.6 MB 182.4 MB 182.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17756 http://ftp.pdbj.org/pub/emdb/structures/EMD-17756 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17756 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17756 | HTTPS FTP |
-Related structure data
| Related structure data |  8pm2MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17756.map.gz / Format: CCP4 / Size: 196.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17756.map.gz / Format: CCP4 / Size: 196.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Consensus map of the mTAAR7f receptor bound to NDMCH in complex with miniGs trimer | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.824 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17756_msk_1.map emd_17756_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Mask #2
| File |  emd_17756_msk_2.map emd_17756_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Receptor-focused map
| File | emd_17756_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Receptor-focused map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_17756_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_17756_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : A complex of mouse trace-amine associated receptor 7f solubilized...
| Entire | Name: A complex of mouse trace-amine associated receptor 7f solubilized in LMNG/CHS bound to N,N-dimethylcyclohexylamine and coupled to: Engineered guanine nucleotide-binding protein G(s) subunit ...Name: A complex of mouse trace-amine associated receptor 7f solubilized in LMNG/CHS bound to N,N-dimethylcyclohexylamine and coupled to: Engineered guanine nucleotide-binding protein G(s) subunit alpha isoform short + Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 + Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 + Nanobody 35 |
|---|---|
| Components |
|
-Supramolecule #1: A complex of mouse trace-amine associated receptor 7f solubilized...
| Supramolecule | Name: A complex of mouse trace-amine associated receptor 7f solubilized in LMNG/CHS bound to N,N-dimethylcyclohexylamine and coupled to: Engineered guanine nucleotide-binding protein G(s) subunit ...Name: A complex of mouse trace-amine associated receptor 7f solubilized in LMNG/CHS bound to N,N-dimethylcyclohexylamine and coupled to: Engineered guanine nucleotide-binding protein G(s) subunit alpha isoform short + Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 + Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 + Nanobody 35 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.964734 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21 (bacteria) Escherichia coli BL21 (bacteria) |
| Sequence | String: GNSKTEDQRN EEKAQREANK KIEKQLQKDK QVYRATHRLL LLGADNSGKS TIVKQMRILH GGSGGSGGTS GIFETKFQVD KVNFHMFDV GGQRDERRKW IQCFNDVTAI IFVVDSSDYN RLQEALNLFK SIWNNRWLRT ISVILFLNKQ DLLAEKVLAG K SKLEDYFP ...String: GNSKTEDQRN EEKAQREANK KIEKQLQKDK QVYRATHRLL LLGADNSGKS TIVKQMRILH GGSGGSGGTS GIFETKFQVD KVNFHMFDV GGQRDERRKW IQCFNDVTAI IFVVDSSDYN RLQEALNLFK SIWNNRWLRT ISVILFLNKQ DLLAEKVLAG K SKLEDYFP EFARYTTPED ATPEPGEDPR VTRAKYFIRD EFLRISTASG DGRHYCYPHF TCAVDTENAR RIFNDCRDII QR MHLRQYE LL |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.342785 KDa |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL ...String: GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD INAICFFPNG NA FATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAGHDNRVSC LGV TDDGMA VATGSWDSFL KIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Details: There is an engineered mutation C68S introduced / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.845078 KDa |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFSAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Nanobody 35
| Macromolecule | Name: Nanobody 35 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Lama glama (llama) Lama glama (llama) |
| Molecular weight | Theoretical: 16.926076 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21 (bacteria) Escherichia coli BL21 (bacteria) |
| Sequence | String: MKYLLPTAAA GLLLLAAQPA MAQVQLQESG GGLVQPGGSL RLSCAASGFT FSNYKMNWVR QAPGKGLEWV SDISQSGASI SYTGSVKGR FTISRDNAKN TLYLQMNSLK PEDTAVYYCA RCPAPFTRDC FDVTSTTYAY RGQGTQVTVS SHHHHHH |
-Macromolecule #5: Trace amine-associated receptor 7f
| Macromolecule | Name: Trace amine-associated receptor 7f / type: protein_or_peptide / ID: 5 Details: mTAAR7f sequence contains cleaved protease sites: TEV (S residue on the N terminus) and HRV-3C (C terminus) Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 41.012539 KDa |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: SMSIADETVS WNQDSILSRD LFSATSAELC YENLNRSCVR SPYSPGPRLI LYAVFGFGAV LAVCGNLLVM TSILHFRQLH SPANFLVAS LACADFLVGV MVMPFSMVRS VEGCWYFGDS YCKLHTCFDV SFCYCSLFHL CFISVDRYIA VSDPLAYPTR F TASVSGKC ...String: SMSIADETVS WNQDSILSRD LFSATSAELC YENLNRSCVR SPYSPGPRLI LYAVFGFGAV LAVCGNLLVM TSILHFRQLH SPANFLVAS LACADFLVGV MVMPFSMVRS VEGCWYFGDS YCKLHTCFDV SFCYCSLFHL CFISVDRYIA VSDPLAYPTR F TASVSGKC ITFSWLLSIS YGFSLIYTGA SEAGLEDLVS SLTCVGGCQI AVNQTWVFIN FSVFLIPTLV MITVYSKIFL IA KQQAQNI EKMSKQTARA SDSYKDRVAK RERKAAKTLG IAVAAFLLSW LPYFIDSFID AFLGFITPTY VYEILVWIVY YNS AMNPLI YAFFYPWFRK AIKLTVTGKI LRENSSTTNL FSELEVLFQ UniProtKB: Trace amine-associated receptor 7f |
-Macromolecule #6: ~{N},~{N}-dimethylcyclohexanamine
| Macromolecule | Name: ~{N},~{N}-dimethylcyclohexanamine / type: ligand / ID: 6 / Number of copies: 1 / Formula: 8IA |
|---|---|
| Molecular weight | Theoretical: 127.227 Da |
| Chemical component information | 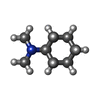 ChemComp-8IA: |
-Macromolecule #7: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 7 / Number of copies: 1 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: GOLD / Support film - topology: HOLEY ARRAY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 120 sec. / Pretreatment - Atmosphere: OTHER Details: Forward Power of 38 W, Reflected Power of 2W; Fischione | ||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 2.4 µm / Calibrated defocus min: 2.4 µm / Calibrated magnification: 96000 / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 0.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 96000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 0.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 96000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 11157 / Average exposure time: 7.84 sec. / Average electron dose: 55.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: Other / Chain - Initial model type: other Details: Initial models of heterotrimeric mini-Gs and Nb35 were sourced from PDB 7T9I. A de novo model of TAAR7f was generated from the focused map and protein sequence using ModelAngelo |
|---|---|
| Refinement | Space: REAL / Protocol: OTHER |
| Output model |  PDB-8pm2: |
 Movie
Movie Controller
Controller
























 Z
Z Y
Y X
X










































