[English] 日本語
 Yorodumi
Yorodumi- EMDB-17724: CryoEM reconstruction of hemagglutinin HK68 of Influenza A virus ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 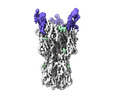 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
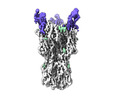
| Title | CryoEM reconstruction of hemagglutinin HK68 of Influenza A virus bound to an Affimer reagent | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Complex / Inhibitor / Complex / Inhibitor /  CryoEM / CryoEM /  Antiviral / Antiviral /  ANTIVIRAL PROTEIN ANTIVIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information viral budding from plasma membrane / clathrin-dependent endocytosis of virus by host cell / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral budding from plasma membrane / clathrin-dependent endocytosis of virus by host cell / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane /  membrane membraneSimilarity search - Function | |||||||||
| Biological species |    Influenza A virus / synthetic construct (others) Influenza A virus / synthetic construct (others) | |||||||||
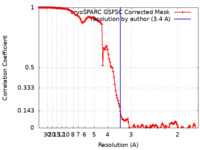
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.4 Å cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Debski-Antoniak O / Flynn A / Klebl DP / Tiede C / Muench S / Tomlinson D / Fontana J | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Exploiting the Affimer platform against influenza A virus Authors: Debski-Antoniak O / Flynn A / Klebl DP / Tiede C / Muench S / Tomlinson D / Fontana J | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17724.map.gz emd_17724.map.gz | 214.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17724-v30.xml emd-17724-v30.xml emd-17724.xml emd-17724.xml | 19.4 KB 19.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17724_fsc.xml emd_17724_fsc.xml | 13.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_17724.png emd_17724.png | 67 KB | ||
| Filedesc metadata |  emd-17724.cif.gz emd-17724.cif.gz | 5.8 KB | ||
| Others |  emd_17724_additional_1.map.gz emd_17724_additional_1.map.gz emd_17724_additional_2.map.gz emd_17724_additional_2.map.gz emd_17724_half_map_1.map.gz emd_17724_half_map_1.map.gz emd_17724_half_map_2.map.gz emd_17724_half_map_2.map.gz | 230.3 MB 121.6 MB 226.5 MB 226.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17724 http://ftp.pdbj.org/pub/emdb/structures/EMD-17724 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17724 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17724 | HTTPS FTP |
-Related structure data
| Related structure data |  8pk3MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17724.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17724.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_17724_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #2
| File | emd_17724_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_17724_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_17724_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : CryoEM reconstruction of hemagglutinin HK68 of Influenza A virus ...
| Entire | Name: CryoEM reconstruction of hemagglutinin HK68 of Influenza A virus bound to an Affimer reagent |
|---|---|
| Components |
|
-Supramolecule #1: CryoEM reconstruction of hemagglutinin HK68 of Influenza A virus ...
| Supramolecule | Name: CryoEM reconstruction of hemagglutinin HK68 of Influenza A virus bound to an Affimer reagent type: complex / ID: 1 / Parent: 0 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:    Influenza A virus Influenza A virus |
-Macromolecule #1: Hemagglutinin HA1 chain
| Macromolecule | Name: Hemagglutinin HA1 chain / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Influenza A virus Influenza A virus |
| Molecular weight | Theoretical: 35.349754 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ADPGATLCLG HHAVPNGTLV KTITDDQIEV TNATELVQSS STGKICNNPH RILDGIDCTL IDALLGDPHC DVFQNETWDL FVERSKAFS NCYPYDVPDY ASLRSLVASS GTLEFITEGF TWTGVTQNGG SNACKRGPGS GFFSRLNWLT KSGSTYPVLN V TMPNNDNF ...String: ADPGATLCLG HHAVPNGTLV KTITDDQIEV TNATELVQSS STGKICNNPH RILDGIDCTL IDALLGDPHC DVFQNETWDL FVERSKAFS NCYPYDVPDY ASLRSLVASS GTLEFITEGF TWTGVTQNGG SNACKRGPGS GFFSRLNWLT KSGSTYPVLN V TMPNNDNF DKLYIWGVHH PSTNQEQTSL YVQASGRVTV STRRSQQTII PNIGSRPWVR GLSSRISIYW TIVKPGDVLV IN SNGNLIA PRGYFKMRTG KSSIMRSDAP IDTCISECIT PNGSIPNDKP FQNVNKITYG ACPKYVKQNT LKLATGMRNV PEK QT UniProtKB:  Hemagglutinin Hemagglutinin |
-Macromolecule #2: Hemagglutinin HA2 chain
| Macromolecule | Name: Hemagglutinin HA2 chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Influenza A virus Influenza A virus |
| Molecular weight | Theoretical: 20.21235 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GLFGAIAGFI ENGWEGMIDG WYGFRHQNSE GTGQAADLKS TQAAIDQING KLNRVIEKTN EKFHQIEKEF SEVEGRIQDL EKYVEDTKI DLWSYNAELL VALENQHTID LTDSEMNKLF EKTRRQLREN AEEMGNGCFK IYHKCDNACI ESIRNGTYDH D VYRDEALN NRFQIKG UniProtKB:  Hemagglutinin Hemagglutinin |
-Macromolecule #3: Affimer molecule (A31)
| Macromolecule | Name: Affimer molecule (A31) / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 10.491819 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: SLEIEELARF AVDEHNKKEN ALLEFVRVVK AKEQFNYDQW QDYTMYYLTL EAKDGGKKKL YEAKVWVKFV DSAFETENFK ELQEFKP |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: OTHER / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.0 µm |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 56.2 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z
Z Y
Y X
X