+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural insights into human TFIIIC promoter recognition | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TFIIIC / tRNA gene / B-Box promoter /  DNA recognition / DNA recognition /  TRANSCRIPTION TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationtRNA transcription / 5S class rRNA transcription by RNA polymerase III / transcription factor TFIIIC complex / RNA polymerase III general transcription initiation factor activity / RNA Polymerase III Transcription Initiation From Type 1 Promoter / RNA Polymerase III Transcription Initiation From Type 2 Promoter / RNA Polymerase III Abortive And Retractive Initiation /  transcription initiation at RNA polymerase III promoter / tRNA transcription by RNA polymerase III / rRNA transcription ...tRNA transcription / 5S class rRNA transcription by RNA polymerase III / transcription factor TFIIIC complex / RNA polymerase III general transcription initiation factor activity / RNA Polymerase III Transcription Initiation From Type 1 Promoter / RNA Polymerase III Transcription Initiation From Type 2 Promoter / RNA Polymerase III Abortive And Retractive Initiation / transcription initiation at RNA polymerase III promoter / tRNA transcription by RNA polymerase III / rRNA transcription ...tRNA transcription / 5S class rRNA transcription by RNA polymerase III / transcription factor TFIIIC complex / RNA polymerase III general transcription initiation factor activity / RNA Polymerase III Transcription Initiation From Type 1 Promoter / RNA Polymerase III Transcription Initiation From Type 2 Promoter / RNA Polymerase III Abortive And Retractive Initiation /  transcription initiation at RNA polymerase III promoter / tRNA transcription by RNA polymerase III / rRNA transcription / transcription initiation at RNA polymerase III promoter / tRNA transcription by RNA polymerase III / rRNA transcription /  enzyme activator activity / transcription by RNA polymerase III / enzyme activator activity / transcription by RNA polymerase III /  histone acetyltransferase activity / histone acetyltransferase activity /  histone acetyltransferase / histone acetyltransferase /  ribonucleoprotein complex / ribonucleoprotein complex /  nucleolus / nucleolus /  mitochondrion / mitochondrion /  DNA binding / DNA binding /  nucleoplasm / nucleoplasm /  membrane membraneSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.4 Å cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Wolfram SD / Mathias G / Luis H / Thomas H / Sebastian E / Christoph M | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Structural insights into human TFIIIC promoter recognition. Authors: Wolfram Seifert-Davila / Mathias Girbig / Luis Hauptmann / Thomas Hoffmann / Sebastian Eustermann / Christoph W Müller /  Abstract: Transcription factor (TF) IIIC recruits RNA polymerase (Pol) III to most of its target genes. Recognition of intragenic A- and B-box motifs in transfer RNA (tRNA) genes by TFIIIC modules τA and τB ...Transcription factor (TF) IIIC recruits RNA polymerase (Pol) III to most of its target genes. Recognition of intragenic A- and B-box motifs in transfer RNA (tRNA) genes by TFIIIC modules τA and τB is the first critical step for tRNA synthesis but is mechanistically poorly understood. Here, we report cryo-electron microscopy structures of the six-subunit human TFIIIC complex unbound and bound to a tRNA gene. The τB module recognizes the B-box via DNA shape and sequence readout through the assembly of multiple winged-helix domains. TFIIIC220 forms an integral part of both τA and τB connecting the two subcomplexes via a ~550-amino acid residue flexible linker. Our data provide a structural mechanism by which high-affinity B-box recognition anchors TFIIIC to promoter DNA and permits scanning for low-affinity A-boxes and TFIIIB for Pol III activation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16717.map.gz emd_16717.map.gz | 248.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16717-v30.xml emd-16717-v30.xml emd-16717.xml emd-16717.xml | 21.6 KB 21.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16717.png emd_16717.png | 96.3 KB | ||
| Filedesc metadata |  emd-16717.cif.gz emd-16717.cif.gz | 8 KB | ||
| Others |  emd_16717_additional_1.map.gz emd_16717_additional_1.map.gz emd_16717_half_map_1.map.gz emd_16717_half_map_1.map.gz emd_16717_half_map_2.map.gz emd_16717_half_map_2.map.gz | 209.7 MB 392.1 MB 392.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16717 http://ftp.pdbj.org/pub/emdb/structures/EMD-16717 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16717 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16717 | HTTPS FTP |
-Related structure data
| Related structure data |  8cllMC  8cliC  8cljC  8clkC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16717.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16717.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.822 Å | ||||||||||||||||||||||||||||||||||||
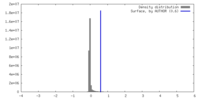
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_16717_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
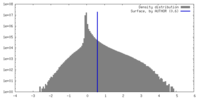
| Density Histograms |
-Half map: #2
| File | emd_16717_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
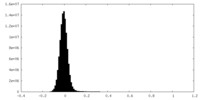
| Density Histograms |
-Half map: #1
| File | emd_16717_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
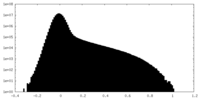
| Density Histograms |
- Sample components
Sample components
-Entire : TFIIIC tauB Dimer
| Entire | Name: TFIIIC tauB Dimer |
|---|---|
| Components |
|
-Supramolecule #1: TFIIIC tauB Dimer
| Supramolecule | Name: TFIIIC tauB Dimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 560 KDa |
-Macromolecule #1: General transcription factor 3C polypeptide 1
| Macromolecule | Name: General transcription factor 3C polypeptide 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 244.200719 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MDALESLLDE VALEGLDGLC LPALWSRLET RVPPFPLPLE PCTQEFLWRA LATHPGISFY EEPRERPDLQ LQDRYEEIDL ETGILESRR DPVALEDVYP IHMILENKDG IQGSCRYFKE RKNITNDIRT KSLQPRCTMV EAFDRWGKKL IIVASQAMRY R ALIGQEGD ...String: MDALESLLDE VALEGLDGLC LPALWSRLET RVPPFPLPLE PCTQEFLWRA LATHPGISFY EEPRERPDLQ LQDRYEEIDL ETGILESRR DPVALEDVYP IHMILENKDG IQGSCRYFKE RKNITNDIRT KSLQPRCTMV EAFDRWGKKL IIVASQAMRY R ALIGQEGD PDLKLPDFSY CILERLGRSR WQGELQRDLH TTAFKVDAGK LHYHRKILNK NGLITMQSHV IRLPTGAQQH SI LLLLNRF HVDRRSKYDI LMEKLSVMLS TRTNHIETLG KLREELGLCE RTFKRLYQYM LNAGLAKVVS LRLQEIHPEC GPC KTKKGT DVMVRCLKLL KEFKRNDHDD DEDEEVISKT VPPVDIVFER DMLTQTYDLI ERRGTKGISQ AEIRVAMNVG KLEA RMLCR LLQRFKVVKG FMEDEGRQRT TKYISCVFAE ESDLSRQYQR EKARSELLTT VSLASMQEES LLPEGEDTFL SESDS EEER SSSKRRGRGS QKDTRASANL RPKTQPHHST PTKGGWKVVN LHPLKKQPPS FPGAAEERAC QSLASRDSLL DTSSVS EPN VSFVSHCADS NSGDIAVIEE VRMENPKESS SSLKTGRHSS GQDKPHETYR LLKRRNLIIE AVTNLRLIES LFTIQKM IM DQEKQEGVST KCCKKSIVRL VRNLSEEGLL RLYRTTVIQD GIKKKVDLVV HPSMDQNDPL VRSAIEQVRF RISNSSTA N RVKTSQPPVP QGEAEEDSQG KEGPSGSGDS QLSASSRSES GRMKKSDNKM GITPLRNYHP IVVPGLGRSL GFLPKMPRL RVVHMFLWYL IYGHPASNTV EKPSFISERR TIKQESGRAG VRPSSSGSAW EACSEAPSKG SQDGVTWEAE VELATETVYV DDASWMRYI PPIPVHRDFG FGWALVSDIL LCLPLSIFIQ IVQVSYKVDN LEEFLNDPLK KHTLIRFLPR PIRQQLLYKR R YIFSVVEN LQRLCYMGLL QFGPTEKFQD KDQVFIFLKK NAVIVDTTIC DPHYNLARSS RPFERRLYVL NSMQDVENYW FD LQCVCLN TPLGVVRCPR VRKNSSTDQG SDEEGSLQKE QESAMDKHNL ERKCAMLEYT TGSREVVDEG LIPGDGLGAA GLD SSFYGH LKRNWIWTSY IINQAKKENT AAENGLTVRL QTFLSKRPMP LSARGNSRLN IWGEARVGSE LCAGWEEQFE VDRE PSLDR NRRVRGGKSQ KRKRLKKDPG KKIKRKKKGE FPGEKSKRLR YHDEADQSAL QRMTRLRVTW SMQEDGLLVL CRIAS NVLN TKVKGPFVTW QVVRDILHAT FEESLDKTSH SVGRRARYIV KNPQAYLNYK VCLAEVYQDK ALVGDFMNRR GDYDDP KVC ANEFKEFVEK LKEKFSSALR NSNLEIPDTL QELFARYRVL AIGDEKDQTR KEDELNSVDD IHFLVLQNLI QSTLALS DS QMKSYQSFQT FRLYREYKDH VLVKAFMECQ KRSLVNRRRV NHTLGPKKNR ALPFVPMSYQ LSQTYYRIFT WRFPSTIC T ESFQFLDRMR AAGKLDQPDR FSFKDQDNNE PTNDMVAFSL DGPGGNCVAV LTLFSLGLIS VDVRIPEQII VVDSSMVEN EVIKSLGKDG SLEDDEDEED DLDEGVGGKR RSMEVKPAQA SHTNYLLMRG YYSPGIVSTR NLNPNDSIVV NSCQMKFQLR CTPVPARLR PAAAPLEELT MGTSCLPDTF TKLINPQENT CSLEEFVLQL ELSGYSPEDL TAALEILEAI IATGCFGIDK E ELRRRFSA LEKAGGGRTR TFADCIQALL EQHQVLEVGG NTARLVAMGS AWPWLLHSVR LKDREDADIQ REDPQARPLE GS SSEDSPP EGQAPPSHSP RGTKRRASWA SENGETDAEG TQMTPAKRPA LQDSNLAPSL GPGAEDGAEA QAPSPPPALE DTA AAGAAQ EDQEGVGEFS SPGQEQLSGQ AQPPEGSEDP RGFTESFGAA NISQAARERD CESVCFIGRP WRVVDGHLNL PVCK GMMEA MLYHIMTRPG IPESSLLRHY QGVLQPVAVL ELLQGLESLG CIRKRWLRKP RPVSLFSTPV VEEVEVPSSL DESPM AFYE PTLDCTLRLG RVFPHEVNWN KWIHLGGGSG GGSGGSLEVL FQGPGSGSDY KDDDDKGDYK DDDDKGDYKD DDDK UniProtKB: General transcription factor 3C polypeptide 1 |
-Macromolecule #2: General transcription factor 3C polypeptide 4
| Macromolecule | Name: General transcription factor 3C polypeptide 4 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO / EC number:  histone acetyltransferase histone acetyltransferase |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 92.093195 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MNTADQARVG PADDGPAPSG EEEGEGGGEA GGKEPAADAA PGPSAAFRLM VTRREPAVKL QYAVSGLEPL AWSEDHRVSV STARSIAVL ELICDVHNPG QDLVIHRTSV PAPLNSCLLK VGSKTEVAEC KEKFAASKDP TVSQTFMLDR VFNPEGKALP P MRGFKYTS ...String: MNTADQARVG PADDGPAPSG EEEGEGGGEA GGKEPAADAA PGPSAAFRLM VTRREPAVKL QYAVSGLEPL AWSEDHRVSV STARSIAVL ELICDVHNPG QDLVIHRTSV PAPLNSCLLK VGSKTEVAEC KEKFAASKDP TVSQTFMLDR VFNPEGKALP P MRGFKYTS WSPMGCDANG RCLLAALTMD NRLTIQANLN RLQWVQLVDL TEIYGERLYE TSYRLSKNEA PEGNLGDFAE FQ RRHSMQT PVRMEWSGIC TTQQVKHNNE CRDVGSVLLA VLFENGNIAV WQFQLPFVGK ESISSCNTIE SGITSPSVLF WWE YEHNNR KMSGLIVGSA FGPIKILPVN LKAVKGYFTL RQPVILWKEM DQLPVHSIKC VPLYHPYQKC SCSLVVAARG SYVF WCLLL ISKAGLNVHN SHVTGLHSLP IVSMTADKQN GTVYTCSSDG KVRQLIPIFT DVALKFEHQL IKLSDVFGSV RTHGI AVSP CGAYLAIITT EGMINGLHPV NKNYQVQFVT LKTFEEAAAQ LLESSVQNLF KQVDLIDLVR WKILKDKHIP QFLQEA LEK KIESSGVTYF WRFKLFLLRI LYQSMQKTPS EALWKPTHED SKILLVDSPG MGNADDEQQE EGTSSKQVVK QGLQERS KE GDVEEPTDDS LPTTGDAGGR EPMEEKLLEI QGKIEAVEMH LTREHMKRVL GEVYLHTWIT ENTSIPTRGL CNFLMSDE E YDDRTARVLI GHISKKMNKQ TFPEHCSLCK EILPFTDRKQ AVCSNGHIWL RCFLTYQSCQ SLIYRRCLLH DSIARHPAP EDPDWIKRLL QSPCPFCDSP VF UniProtKB: General transcription factor 3C polypeptide 4 |
-Macromolecule #3: General transcription factor 3C polypeptide 2
| Macromolecule | Name: General transcription factor 3C polypeptide 2 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 102.612453 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MHHHHHHENL YFQGMDTCGV GYVALGEAGP VGNMTVVDSP GQEVLNQLDV KTSSEMTSAE ASVEMSLPTP LPGFEDSPDQ RRLPPEQES LSRLEQPDLS SEMSKVSKPR ASKPGRKRGG RTRKGPKRPQ QPNPPSAPLV PGLLDQSNPL STPMPKKRGR K SKAELLLL ...String: MHHHHHHENL YFQGMDTCGV GYVALGEAGP VGNMTVVDSP GQEVLNQLDV KTSSEMTSAE ASVEMSLPTP LPGFEDSPDQ RRLPPEQES LSRLEQPDLS SEMSKVSKPR ASKPGRKRGG RTRKGPKRPQ QPNPPSAPLV PGLLDQSNPL STPMPKKRGR K SKAELLLL KLSKDLDRPE SQSPKRPPED FETPSGERPR RRAAQVALLY LQELAEELST ALPAPVSCPE GPKVSSPTKP KK IRQPAAC PGGEEVDGAP RDEDFFLQVE AEDVEESEGP SESSSEPEPV VPRSTPRGST SGKQKPHCRG MAPNGLPNHI MAP VWKCLH LTKDFREQKH SYWEFAEWIP LAWKWHLLSE LEAAPYLPQE EKSPLFSVQR EGLPEDGTLY RINRFSSITA HPER WDVSF FTGGPLWALD WCPVPEGAGA SQYVALFSSP DMNETHPLSQ LHSGPGLLQL WGLGTLQQES CPGNRAHFVY GIACD NGCI WDLKFCPSGA WELPGTPRKA PLLPRLGLLA LACSDGKVLL FSLPHPEALL AQQPPDAVKP AIYKVQCVAT LQVGSM QAT DPSECGQCLS LAWMPTRPHQ HLAAGYYNGM VVFWNLPTNS PLQRIRLSDG SLKLYPFQCF LAHDQAVRTL QWCKANS HF LVSAGSDRKI KFWDLRRPYE PINSIKRFLS TELAWLLPYN GVTVAQDNCY ASYGLCGIHY IDAGYLGFKA YFTAPRKG T VWSLSGSDWL GTIAAGDISG ELIAAILPDM ALNPINVKRP VERRFPIYKA DLIPYQDSPE GPDHSSASSG VPNPPKART YTETVNHHYL LFQDTDLGSF HDLLRREPML RMQEGEGHSQ LCLDRLQLEA IHKVRFSPNL DSYGWLVSGG QSGLVRIHFV RGLASPLGH RMQLESRAHF NAMFQPSSPT RRPGFSPTSH RLLPTP UniProtKB: General transcription factor 3C polypeptide 2 |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.7 µm / Nominal defocus min: 0.7000000000000001 µm Bright-field microscopy / Nominal defocus max: 1.7 µm / Nominal defocus min: 0.7000000000000001 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 42.8 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL / In silico model: Alphafold2 model |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 125600 |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)