[English] 日本語
 Yorodumi
Yorodumi- EMDB-16657: Cryo-EM structure of the fd bacteriophage capsid major coat prote... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the fd bacteriophage capsid major coat protein pVIII | ||||||||||||||||||||||||
 Map data Map data | Cryo-EM map of the intact fd bacteriophage capsid | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords |  Bacteriophage / fd / Bacteriophage / fd /  inovirus / ff / helical / inovirus / ff / helical /  phage / phage /  filamentous / filamentous /  VIRUS VIRUS | ||||||||||||||||||||||||
| Function / homology | Phage major coat protein, Gp8 /  Bacteriophage M13, G8P, capsid domain superfamily / Bacteriophage M13, G8P, capsid domain superfamily /  Capsid protein G8P / helical viral capsid / host cell membrane / Capsid protein G8P / helical viral capsid / host cell membrane /  membrane / membrane /  Capsid protein G8P Capsid protein G8P Function and homology information Function and homology information | ||||||||||||||||||||||||
| Biological species |   Escherichia coli (E. coli) / Escherichia coli (E. coli) /  Enterobacteria phage fd (virus) Enterobacteria phage fd (virus) | ||||||||||||||||||||||||
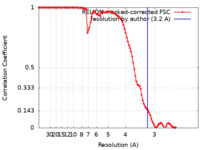
| Method | helical reconstruction /  cryo EM / Resolution: 3.2 Å cryo EM / Resolution: 3.2 Å | ||||||||||||||||||||||||
 Authors Authors | Boehning J / Bharat TAM | ||||||||||||||||||||||||
| Funding support |  United Kingdom, United Kingdom,  France, France,  United States, European Union, 7 items United States, European Union, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Biophysical basis of filamentous phage tactoid-mediated antibiotic tolerance in P. aeruginosa. Authors: Jan Böhning / Miles Graham / Suzanne C Letham / Luke K Davis / Ulrike Schulze / Phillip J Stansfeld / Robin A Corey / Philip Pearce / Abul K Tarafder / Tanmay A M Bharat /  Abstract: Inoviruses are filamentous phages infecting numerous prokaryotic phyla. Inoviruses can self-assemble into mesoscale structures with liquid-crystalline order, termed tactoids, which protect bacterial ...Inoviruses are filamentous phages infecting numerous prokaryotic phyla. Inoviruses can self-assemble into mesoscale structures with liquid-crystalline order, termed tactoids, which protect bacterial cells in Pseudomonas aeruginosa biofilms from antibiotics. Here, we investigate the structural, biophysical, and protective properties of tactoids formed by the P. aeruginosa phage Pf4 and Escherichia coli phage fd. A cryo-EM structure of the capsid from fd revealed distinct biochemical properties compared to Pf4. Fd and Pf4 formed tactoids with different morphologies that arise from differing phage geometries and packing densities, which in turn gave rise to different tactoid emergent properties. Finally, we showed that tactoids formed by either phage protect rod-shaped bacteria from antibiotic treatment, and that direct association with a tactoid is required for protection, demonstrating the formation of a diffusion barrier by the tactoid. This study provides insights into how filamentous molecules protect bacteria from extraneous substances in biofilms and in host-associated infections. #1:  Journal: Biorxiv / Year: 2023 Journal: Biorxiv / Year: 2023Title: Biophysical basis of phage liquid crystalline droplet-mediated antibiotic tolerance in pathogenic bacteria Authors: Bohning J / Graham M / Letham S / Davis L / Schulze U / Stansfeld P / Corey R / Pearce P / Tarafder A / Bharat T | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16657.map.gz emd_16657.map.gz | 96.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16657-v30.xml emd-16657-v30.xml emd-16657.xml emd-16657.xml | 18.8 KB 18.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16657_fsc.xml emd_16657_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_16657.png emd_16657.png | 101.5 KB | ||
| Filedesc metadata |  emd-16657.cif.gz emd-16657.cif.gz | 5.9 KB | ||
| Others |  emd_16657_half_map_1.map.gz emd_16657_half_map_1.map.gz emd_16657_half_map_2.map.gz emd_16657_half_map_2.map.gz | 81.1 MB 80.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16657 http://ftp.pdbj.org/pub/emdb/structures/EMD-16657 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16657 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16657 | HTTPS FTP |
-Related structure data
| Related structure data |  8ch5MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16657.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16657.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of the intact fd bacteriophage capsid | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.092 Å | ||||||||||||||||||||||||||||||||||||
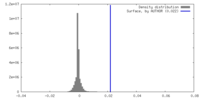
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half-map 2
| File | emd_16657_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
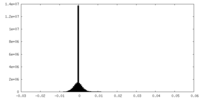
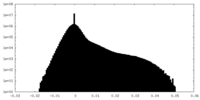
| Density Histograms |
-Half map: Half-map 1
| File | emd_16657_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
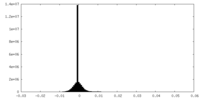
| Density Histograms |
- Sample components
Sample components
-Entire : Enterobacteria phage fd
| Entire | Name:  Enterobacteria phage fd (virus) Enterobacteria phage fd (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Enterobacteria phage fd
| Supramolecule | Name: Enterobacteria phage fd / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Purified by PEG precipitation from the supernatant of infected E. coli cells. NCBI-ID: 10864 / Sci species name: Enterobacteria phage fd / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Macromolecule #1: Major capsid protein pVIII
| Macromolecule | Name: Major capsid protein pVIII / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 5.244 KDa |
| Sequence | String: AEGDDPAKAA FDSLQASATE YIGYAWAMVV VIVGATIGIK LFKKFTSKAS UniProtKB:  Capsid protein G8P Capsid protein G8P |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: Phosphate-buffered saline |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER/RHODIUM / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: AIR / Details: 15 mA |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
| Details | fd phage in PBS |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average electron dose: 53.9 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 79.94 |
|---|---|
| Output model |  PDB-8ch5: |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)