+ Open data
Open data
- Basic information
Basic information
| Entry | 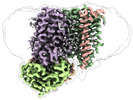 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
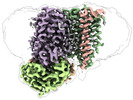
| Title | Cytochrome c maturation complex CcmABCD, E154Q, ATP-bound | |||||||||||||||
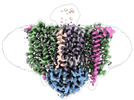
 Map data Map data | sharpened map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Cytochrome c maturation /  MEMBRANE PROTEIN MEMBRANE PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcytochrome c biosynthetic process / heme import across plasma membrane / ABC-type heme transporter / ABC-type heme transporter activity / heme transmembrane transporter activity / cytochrome complex assembly / ATP-binding cassette (ABC) transporter complex /  heme binding / heme binding /  ATP hydrolysis activity / ATP hydrolysis activity /  ATP binding / ATP binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||||||||
| Biological species |   Escherichia coli (E. coli) / Escherichia coli (E. coli) /   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria) | |||||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.62 Å cryo EM / Resolution: 3.62 Å | |||||||||||||||
 Authors Authors | Ilcu L / Zhang L / Einsle O | |||||||||||||||
| Funding support | European Union,  Germany, 4 items Germany, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Architecture of the Heme-translocating CcmABCD/E complex required for Cytochrome c maturation. Authors: Lorena Ilcu / Lukas Denkhaus / Anton Brausemann / Lin Zhang / Oliver Einsle /  Abstract: Mono- and multiheme cytochromes c are post-translationally matured by the covalent attachment of heme. For this, Escherichia coli employs the most complex type of maturation machineries, the Ccm- ...Mono- and multiheme cytochromes c are post-translationally matured by the covalent attachment of heme. For this, Escherichia coli employs the most complex type of maturation machineries, the Ccm-system (for cytochrome c maturation). It consists of two membrane protein complexes, one of which shuttles heme across the membrane to a mobile chaperone that then delivers the cofactor to the second complex, an apoprotein:heme lyase, for covalent attachment. Here we report cryo-electron microscopic structures of the heme translocation complex CcmABCD from E. coli, alone and bound to the heme chaperone CcmE. CcmABCD forms a heterooctameric complex centered around the ABC transporter CcmAB that does not by itself transport heme. Our data suggest that the complex flops a heme group from the inner to the outer leaflet at its CcmBC interfaces, driven by ATP hydrolysis at CcmA. A conserved heme-handling motif (WxWD) at the periplasmic side of CcmC rotates the heme by 90° for covalent attachment to the heme chaperone CcmE that we find interacting exclusively with the CcmB subunit. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16599.map.gz emd_16599.map.gz | 48.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16599-v30.xml emd-16599-v30.xml emd-16599.xml emd-16599.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16599.png emd_16599.png | 124.2 KB | ||
| Others |  emd_16599_additional_1.map.gz emd_16599_additional_1.map.gz emd_16599_half_map_1.map.gz emd_16599_half_map_1.map.gz emd_16599_half_map_2.map.gz emd_16599_half_map_2.map.gz | 47.1 MB 48.8 MB 48.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16599 http://ftp.pdbj.org/pub/emdb/structures/EMD-16599 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16599 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16599 | HTTPS FTP |
-Related structure data
| Related structure data |  8ce5MC  8ce1C  8ce8C  8ceaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16599.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16599.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.878 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: unsharpened map
| File | emd_16599_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
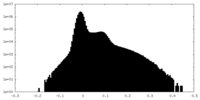
| Density Histograms |
-Half map: half map A
| File | emd_16599_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A
| File | emd_16599_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of cytochrome c maturation sysmtem I, Ccm(AB)2CD
| Entire | Name: Complex of cytochrome c maturation sysmtem I, Ccm(AB)2CD |
|---|---|
| Components |
|
-Supramolecule #1: Complex of cytochrome c maturation sysmtem I, Ccm(AB)2CD
| Supramolecule | Name: Complex of cytochrome c maturation sysmtem I, Ccm(AB)2CD type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Macromolecule #1: Cytochrome c biogenesis ATP-binding export protein CcmA
| Macromolecule | Name: Cytochrome c biogenesis ATP-binding export protein CcmA type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: ABC-type heme transporter |
|---|---|
| Source (natural) | Organism:   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria) |
| Molecular weight | Theoretical: 24.410729 KDa |
| Recombinant expression | Organism:   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria) |
| Sequence | String: MASWSHPQFE KMGMLEAREL LCERDERTLF SGLSFTLNAG EWVQITGSNG AGKTTLLRLL TGLSRPDAGE VLWQGQPLHQ VRDSYHQNL LWIGHQPGIK TRLTALENLH FYHRDGDTAQ CLEALAQAGL AGFEDIPVNQ LSAGQQRRVA LARLWLTRAT L WILDQPFT ...String: MASWSHPQFE KMGMLEAREL LCERDERTLF SGLSFTLNAG EWVQITGSNG AGKTTLLRLL TGLSRPDAGE VLWQGQPLHQ VRDSYHQNL LWIGHQPGIK TRLTALENLH FYHRDGDTAQ CLEALAQAGL AGFEDIPVNQ LSAGQQRRVA LARLWLTRAT L WILDQPFT AIDVNGVDRL TQRMAQHTEQ GGIVILTTHQ PLNVAESKIR RISLTQTRAA UniProtKB: Cytochrome c biogenesis ATP-binding export protein CcmA |
-Macromolecule #2: Heme exporter protein B
| Macromolecule | Name: Heme exporter protein B / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria) |
| Molecular weight | Theoretical: 23.632676 KDa |
| Recombinant expression | Organism:   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria) |
| Sequence | String: MMFWRIFRLE LRVAFRHSAE IANPLWFFLI VITLFPLSIG PEPQLLARIA PGIIWVAALL SSLLALERLF RDDLQDGSLE QLMLLPLPL PAVVLAKVMA HWMVTGLPLL ILSPLVAMLL GMDVYGWQVM ALTLLLGTPT LGFLGAPGVA LTVGLKRGGV L LSILVLPL ...String: MMFWRIFRLE LRVAFRHSAE IANPLWFFLI VITLFPLSIG PEPQLLARIA PGIIWVAALL SSLLALERLF RDDLQDGSLE QLMLLPLPL PAVVLAKVMA HWMVTGLPLL ILSPLVAMLL GMDVYGWQVM ALTLLLGTPT LGFLGAPGVA LTVGLKRGGV L LSILVLPL TIPLLIFATA AMDAASMHLP VDGYLAILGA LLAGTATLSP FATAAALRIS IQ UniProtKB: Heme exporter protein B |
-Macromolecule #3: Heme exporter protein C
| Macromolecule | Name: Heme exporter protein C / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria) |
| Molecular weight | Theoretical: 27.911264 KDa |
| Recombinant expression | Organism:   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria) |
| Sequence | String: MWKTLHQLAI PPRLYQICGW FIPWLAIASV VVLTVGWIWG FGFAPADYQQ GNSYRIIYLH VPAAIWSMGI YASMAVAAFI GLVWQMKMA NLAVAAMAPI GAVFTFIALV TGSAWGKPMW GTWWVWDARL TSELVLLFLY VGVIALWHAF DDRRLAGRAA G ILVLIGVV ...String: MWKTLHQLAI PPRLYQICGW FIPWLAIASV VVLTVGWIWG FGFAPADYQQ GNSYRIIYLH VPAAIWSMGI YASMAVAAFI GLVWQMKMA NLAVAAMAPI GAVFTFIALV TGSAWGKPMW GTWWVWDARL TSELVLLFLY VGVIALWHAF DDRRLAGRAA G ILVLIGVV NLPIIHYSVE WWNTLHQGST RMQQSIDPAM RSPLRWSIFG FLLLSATLTL MRMRNLILLM EKRRPWVSEL IL KRGRK UniProtKB: Heme exporter protein C |
-Macromolecule #4: Heme exporter protein D
| Macromolecule | Name: Heme exporter protein D / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria) |
| Molecular weight | Theoretical: 7.753103 KDa |
| Recombinant expression | Organism:   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria) |
| Sequence | String: MTPAFASWNE FFAMGGYAFF VWLAVVMTVI PLVVLVVHSV MQHRAILRGV AQQRAREARL RAAQQQEAA UniProtKB: Heme exporter protein D |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 6 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.62 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 355914 |
 Movie
Movie Controller
Controller



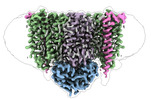



 Z
Z Y
Y X
X