+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Slipper limpet hemocyanin tridecamer | ||||||||||||
 Map data Map data | Slipper limpet hemocyanin tridecamer | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords |  Hemocyanin / tridecamer / copper-oxygen / Hemocyanin / tridecamer / copper-oxygen /  OXYGEN TRANSPORT OXYGEN TRANSPORT | ||||||||||||
| Biological species |  Crepidula fornicata (invertebrata) Crepidula fornicata (invertebrata) | ||||||||||||
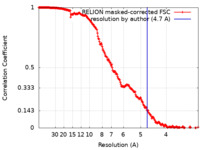
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.7 Å cryo EM / Resolution: 4.7 Å | ||||||||||||
 Authors Authors | Clare D / Young MT | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: PLoS One / Year: 2023 Journal: PLoS One / Year: 2023Title: CryoEM structure and Alphafold molecular modelling of a novel molluscan hemocyanin. Authors: Gaia Pasqualetto / Andrew Mack / Emily Lewis / Ryan Cooper / Alistair Holland / Ufuk Borucu / Judith Mantell / Tom Davies / Miriam Weckener / Dan Clare / Tom Green / Pete Kille / Alex Muhlhozl / Mark T Young /  Abstract: Hemocyanins are multimeric oxygen transport proteins present in the blood of arthropods and molluscs, containing up to 8 oxygen-binding functional units per monomer. In molluscs, hemocyanins are ...Hemocyanins are multimeric oxygen transport proteins present in the blood of arthropods and molluscs, containing up to 8 oxygen-binding functional units per monomer. In molluscs, hemocyanins are assembled in decamer 'building blocks' formed of 5 dimer 'plates', routinely forming didecamer or higher-order assemblies with d5 or c5 symmetry. Here we describe the cryoEM structures of the didecamer (20-mer) and tridecamer (30-mer) forms of a novel hemocyanin from the slipper limpet Crepidula fornicata (SLH) at 7.0 and 4.7 Å resolution respectively. We show that two decamers assemble in a 'tail-tail' configuration, forming a partially capped cylinder, with an additional decamer adding on in 'head-tail' configuration to make the tridecamer. Analysis of SLH samples shows substantial heterogeneity, suggesting the presence of many higher-order multimers including tetra- and pentadecamers, formed by successive addition of decamers in head-tail configuration. Retrieval of sequence data for a full-length isoform of SLH enabled the use of Alphafold to produce a molecular model of SLH, which indicated the formation of dimer slabs with high similarity to those found in keyhole limpet hemocyanin. The fit of the molecular model to the cryoEM density was excellent, showing an overall structure where the final two functional units of the subunit (FU-g and FU-h) form the partial cap at one end of the decamer, and permitting analysis of the subunit interfaces governing the assembly of tail-tail and head-tail decamer interactions as well as potential sites for N-glycosylation. Our work contributes to the understanding of higher-order oligomer formation in molluscan hemocyanins and demonstrates the utility of Alphafold for building accurate structural models of large oligomeric proteins. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16523.map.gz emd_16523.map.gz | 74.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16523-v30.xml emd-16523-v30.xml emd-16523.xml emd-16523.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16523_fsc.xml emd_16523_fsc.xml | 21.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_16523.png emd_16523.png | 93.4 KB | ||
| Masks |  emd_16523_msk_1.map emd_16523_msk_1.map | 824 MB |  Mask map Mask map | |
| Others |  emd_16523_half_map_1.map.gz emd_16523_half_map_1.map.gz emd_16523_half_map_2.map.gz emd_16523_half_map_2.map.gz | 672.4 MB 672.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16523 http://ftp.pdbj.org/pub/emdb/structures/EMD-16523 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16523 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16523 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16523.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16523.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Slipper limpet hemocyanin tridecamer | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.60018 Å | ||||||||||||||||||||||||||||||||||||
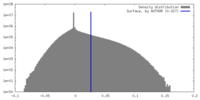
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16523_msk_1.map emd_16523_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
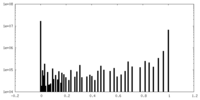
| Density Histograms |
-Half map: #2
| File | emd_16523_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
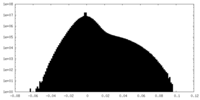
| Density Histograms |
-Half map: #1
| File | emd_16523_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Slipper limpet hemocyanin
| Entire | Name: Slipper limpet hemocyanin |
|---|---|
| Components |
|
-Supramolecule #1: Slipper limpet hemocyanin
| Supramolecule | Name: Slipper limpet hemocyanin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Crepidula fornicata (invertebrata) Crepidula fornicata (invertebrata) |
| Molecular weight | Theoretical: 10.5 MDa |
-Macromolecule #1: Slipper limpet hemocyanin
| Macromolecule | Name: Slipper limpet hemocyanin / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Crepidula fornicata (invertebrata) Crepidula fornicata (invertebrata) |
| Sequence | String: MTTFCGPLAL LVLWCQLLLA SGSLIRKNVD SLTESEVLTL QTVLRQLEDD KTATGFQAIA AYHGEPSQCQ TKDGTAIACC VHGEATFPQW HRLYVVQFEQ ALVSKGLANI GVPYWDWTKP IKDLPALVKN QIFRDPKGKL GQKNAWFSAE IDHDNVHTQT ARAIDDRLYE ...String: MTTFCGPLAL LVLWCQLLLA SGSLIRKNVD SLTESEVLTL QTVLRQLEDD KTATGFQAIA AYHGEPSQCQ TKDGTAIACC VHGEATFPQW HRLYVVQFEQ ALVSKGLANI GVPYWDWTKP IKDLPALVKN QIFRDPKGKL GQKNAWFSAE IDHDNVHTQT ARAIDDRLYE SVSPGGHTTL FNMVLDALEQ DDYCQFEAQF EVAHNHIHYL VGGRHHYSMS TLEYTSFDPL FFLHHSNVDR IFAIWQELQK KRGKPYDHAD CSVELFNKDL LPFARDSNPI QLTRTFSKPK DLFSYMQLGY TYDDLTLNDM SIDQLYSFLK ERHARERSFA VFSLHGMGFS ANVKVSVCNV EEEETTNCEF AGDIFILGGA NEMAWEFTVP YYYDISDAVR KLGHQQDFNF GVKAEVYSVN GTHLGSDVLS RPYASHRNPR GYTDPVIRDS RKPSNVVRKD IELLTDEEVY ALRQAMHRFQ NDTSIDGYQA VAEFHGLPAK CPHPDAAVRY ACCVHGMATF PHWHRLFVVE VEDELRSRGL EIGIPYWDWT RPNAHIPALA NDVSYEDANT HQQLHNPFHD APIAFLGEKT TRQVQDDLSE SPEFGDHTSL FDGILLAFEQ TDFCDFEVQF EVVHNAPHFL VGGFGAYTLA TLHYSAFDPI FYLHHSNVDR LWAIWQQLQI KRGLPYKAHC ASSLTQEVLK PFGFPPPWNN DHKTFENARP TNIYDYEAVL DYTYDSLQFG GMTVDQLEHY LHERKTQNRD FVGIALHNIG VSAWVTLSLQ VEGGEPYQVG KIAVLGGEKE MPWRFDRLFK VEITSALQKL GLSYDDDFNV HLDITDVTGK KWAEDTFHHN TIIHVPGEGH EDPGEDDDHV RNDIVTLTAE QEQNVRDAFQ GLKNDKSVAG YNQIAAFHGQ PNWCPSTQAE RKYACCTHGM AVFPHWHRLL TVQAENGLIA NGLHSGLPYW DWTLPMTSLP SIVLNATYVH PKTGESVANP LYSGQVDGHD TTRSVRQELF EQPKFGHMTK IAEKVMLAFE QDNFCDFEIQ YEIAHNYIHA LVGGNETYSM ASLRYTAYDP IFFLHHSNTD RIWAIWQALQ KYRGKPYNSA NCAIAEMRKP LQPFAQTSLT NPDYVTRDHS TPFDVFNYHS SFHYHYDNLD FNGMSVAQLQ REVIRRRGLE RAFAGFMLHG VKKSCLVVFD ICKPDGTCTK SGEFYLLGDE NELPWEYDRL YKYEITHELE DMGLEPQDRF DVQYHVYDLD NTDLGDDVFG KAVIIYSAGQ GHQKGHEEDY LEEVQASSHV RRNLEDLTTG EAESLRSALR DMEEDGSFDA IARFHGYPGL CEHDGHKAAC CVHGSPAFPH WHRLYVEQVE NSLLSHGSAV SVPYWDWTQP IKKLPALIND ATYYDSRAHA KLENPFFRWK IPGTDMYTSR DPRPDLFDSD YFLNNALLAL EQTSYCDFEV QFEILHNALH SFLGGRGKYS LSSLDYSAFD PVFFLHHANV DRIWAIWQAL QKIRGLPFDE SDCALNIMGT PLHPFDDKEE NQFDLTNKYS RPIDAFDYSN HFDYHYDTLK FNSWTIPQLE QVLEKQRSRD RVFAGFLLHN IGTSADVEID VCVATGNGAK SCNHPAGKFA ILGGEYEMPF TFDRLYKYDI SDTVRKLGLR LDSAADFDVQ IKIFAYNGSY IDASLLHRPT IIFEPGQGQT QDDVGHVERR LVRKSVLSLS PAERRSLVLA MRSLQEDSSA DGFQSLASFH ALPPLCPYPE ATKRFACCIH GMATFPHWHR LYTVQFEDAL RRHGALVGIP YWDTVVPNSR LPDFIAESVW DDPLFHANFS NPWAGADIEF DNSAVVRDVN MDRISQKGPK GYDTWSWKQY LYALEQENFC DFEVQFEIAH NALHAWMGGS EVHSMAHLHY ASYDPVFILH HSNTDRIFAL WQELQKYRGR DANEVNCALE LMKEPLKPFS FGSPYNLNPT TTLYSKPEDA FDYKGHFKYE YDTLELQGLD VQRLQDYINK EKEHDRVFAG FLLSGIGQSA HASFSVCKAN GECTAAGDFD ILGGSAEMPW RFDRLYKYEI TDVLEKKGLD VHDSFNITVS LTALDGSALS SSLLPTPSVI FEPKSRTTEL HEVAPNRIRH DLTHLSERDI RSLKSSIRDL QLDDSNDGYQ NIASFHGAPA LCPSPEAAEY ACCVHGMPTF PHWHRLYTVE MEDAMVRHGS SVALPYWDWT MPITQLPDLF TSETYYDAWR DEVVANPFAR AYIKVAGGYT VRDPQAQLKQ LSRDGQHSAL FDLVLLVLEQ TDYCDFEVQF EVVHNAIHYL VGGRQLYSMS SLEYTSYDPI FFVHHSFVDK IWAVWQELQK RRGMNYDRAD CAVNFMNERM HPFDWEALNP DVRTREHSLP QSVWKYEDLG YHYDNYQIGG KSIEELEELI KKQQSHPRVF AGFQLHGLGT SADVELSVCK SQNSCVSAGV IFILGGKLEM PWAFDRLFKL DITDTLHDMG IEPEDVFDTQ APFFLSYEVH AVNGTTLPLS TISPPTLVFQ PAEGAAAEHS SYSIAGVGVR KDINTLTAAE MENLRDALGR VQAGTGRLTY DNVVSAHGYP PQCTHDGHKV ACCQHGMASF PMWHRLFTRQ MEVALSWEGA KVGIPYWDWT EAFTELPALV REEENNPFHH GRIPGTDTVT TRAPRPQLFR DPEHGDESFF YRQVQLALEQ RDYCDFEVQF EVIHNAIHSW IGGTSPYGMS TLEYSAYDPV FFIHHSNVDR QFAIWQALQK YRGLDYNTAN CNIQELRQVQ EPFDRDDNPV VTTRHYSKAI DAFNYDQYGY QYDNLNFHGM TISQLDEMLE KKKQEDHVFA NFMLHGIQTS ADVVFDLCDA KGKCNFAGTF AILGGPLEMP WSFDRLFKYD VTSVFKQMRL RPDSEYSFRV SLTAVNGTQL DSRLIEAPSV SFVPGNKPGS KTSAAHEDPV PLDTGDVTRY EVSALSLAQV TNLRDALYKL QNDHGPNGFE AIASFHGAPG LCPENATDHY ACCRHGMPAF PHWHRLLTVQ FERALKDKGA VVGVPYWDWT RPAKAMPSLF TDSYDNNPFQ TYRMTFNDQY IQRDVSEELF NHPSEGDVES LFHQALETLE ETNYCDFEVQ YEMLHNAVHE LIGGGNTYSM STLEYSAFDP FFMVHHASID RIWQIWQSLQ KLRHKPFNYA SCASRSLYKP LEPFSYTSLN ADPLTLNNAQ PVHIFDTAKF HYHYDSLDLN GHGVHELHDM IEDMQATSRI FAGFVLSGIS TSARVHVDVT RGEDTVSVGN FYVLGGSSEM PWAYERIYKL DMTEAASKLG LSSESTFHFK LTVTKYDGTA LNVTFPDPVI VKRAANSQHD VLVLPLSVAN QLPPKIVVRQ GTQVVFHASE SGVSSLREVG SYTNSVHCAI PPGQANLYDL DVAYSLEAGD YYFTSSDKTK CQQGSRIQIT VDDE |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 15 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: 50 mM Tris-HCl, 150 mM NaCl, 5 mM MgCl2, 5 mM CaCl2 |
| Grid | Mesh: 300 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 6.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 92000 Bright-field microscopy / Nominal defocus max: 6.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 92000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 856 / Average exposure time: 13.0 sec. / Average electron dose: 2.23 e/Å2 |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)