[English] 日本語
 Yorodumi
Yorodumi- EMDB-16009: Hepatitis B virus core antigen (HBc) with the insertion of four e... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Hepatitis B virus core antigen (HBc) with the insertion of four external domains of the influenza A M2 protein (HBc/4M2e) with T=3 topology | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationsuppression by virus of host autophagy / microtubule-dependent intracellular transport of viral material towards nucleus / T=4 icosahedral viral capsid / proton transmembrane transporter activity / virus-mediated perturbation of host defense response / viral penetration into host nucleus / : / protein complex oligomerization / monoatomic ion channel activity / host cell cytoplasm ...suppression by virus of host autophagy / microtubule-dependent intracellular transport of viral material towards nucleus / T=4 icosahedral viral capsid / proton transmembrane transporter activity / virus-mediated perturbation of host defense response / viral penetration into host nucleus / : / protein complex oligomerization / monoatomic ion channel activity / host cell cytoplasm / symbiont entry into host cell / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity /  DNA binding / DNA binding /  RNA binding / extracellular region / RNA binding / extracellular region /  membrane membraneSimilarity search - Function | |||||||||
| Biological species |    Hepatitis B virus Hepatitis B virus | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.0 Å cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Egorov VV / Shvetsov AV / Pichkur EB / Shaldzhyan AA / Zabrodskaya YA / Vinogradova DS / Nekrasov PA / Gorshkov AN / Garmay YP / Kovaleva AA ...Egorov VV / Shvetsov AV / Pichkur EB / Shaldzhyan AA / Zabrodskaya YA / Vinogradova DS / Nekrasov PA / Gorshkov AN / Garmay YP / Kovaleva AA / Stepanova LA / Tsybalova LM / Shtam TA / Myasnikov AG / Konevega AL | |||||||||
| Funding support |  Russian Federation, 1 items Russian Federation, 1 items
| |||||||||
 Citation Citation |  Journal: Biophys Chem / Year: 2023 Journal: Biophys Chem / Year: 2023Title: Inside and outside of virus-like particles HBc and HBc/4M2e: A comprehensive study of the structure. Authors: V V Egorov / A V Shvetsov / E B Pichkur / A A Shaldzhyan / Ya A Zabrodskaya / D S Vinogradova / P A Nekrasov / A N Gorshkov / Yu P Garmay / A A Kovaleva / L A Stepanova / L M Tsybalova / T A ...Authors: V V Egorov / A V Shvetsov / E B Pichkur / A A Shaldzhyan / Ya A Zabrodskaya / D S Vinogradova / P A Nekrasov / A N Gorshkov / Yu P Garmay / A A Kovaleva / L A Stepanova / L M Tsybalova / T A Shtam / A G Myasnikov / A L Konevega /  Abstract: Hepatitis B virus core antigen (HBc) with the insertion of four external domains of the influenza A M2 protein (HBc/4M2e) form virus-like particles whose structure was studied using a combination of ...Hepatitis B virus core antigen (HBc) with the insertion of four external domains of the influenza A M2 protein (HBc/4M2e) form virus-like particles whose structure was studied using a combination of molecular modeling and cryo-electron microscopy (cryo-EM). It was also shown that self-assembling of the particles occurs inside bacterial cells, but despite the big inner volume of the core shell particle, purified HBc/4M2e contain an insignificant amount of bacterial proteins. It was shown that a fragment of the M2e corresponding to 4M2e insertion is prone to formation of amyloid-like fibrils. However, as the part of the immunodominant loop, M2e insertion does not show a tendency to intermolecular interaction. A full-atomic HBc-4M2e model with the resolution of about 3 Å (3.13 Å for particles of Т = 4 symmetry, 3.7 Å for particles of Т = 3 symmetry) was obtained by molecular modeling methods based on cryo-EM data. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16009.map.gz emd_16009.map.gz | 306.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16009-v30.xml emd-16009-v30.xml emd-16009.xml emd-16009.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16009.png emd_16009.png | 135.8 KB | ||
| Masks |  emd_16009_msk_1.map emd_16009_msk_1.map | 325 MB |  Mask map Mask map | |
| Others |  emd_16009_half_map_1.map.gz emd_16009_half_map_1.map.gz emd_16009_half_map_2.map.gz emd_16009_half_map_2.map.gz | 296.9 MB 297 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16009 http://ftp.pdbj.org/pub/emdb/structures/EMD-16009 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16009 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16009 | HTTPS FTP |
-Related structure data
| Related structure data |  8berMC  8bdzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16009.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16009.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.107 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16009_msk_1.map emd_16009_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
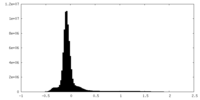
| Density Histograms |
-Half map: #2
| File | emd_16009_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
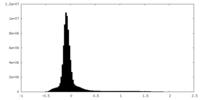
| Density Histograms |
-Half map: #1
| File | emd_16009_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
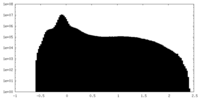
| Density Histograms |
- Sample components
Sample components
-Entire : Hepatitis B virus core antigen (HBc) with the insertion of four e...
| Entire | Name: Hepatitis B virus core antigen (HBc) with the insertion of four external domains of the influenza A M2 protein (T=3) |
|---|---|
| Components |
|
-Supramolecule #1: Hepatitis B virus core antigen (HBc) with the insertion of four e...
| Supramolecule | Name: Hepatitis B virus core antigen (HBc) with the insertion of four external domains of the influenza A M2 protein (T=3) type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:    Hepatitis B virus Hepatitis B virus |
| Molecular weight | Theoretical: 30.84 kDa/nm |
-Macromolecule #1: Core protein,Matrix protein 2,External core antigen
| Macromolecule | Name: Core protein,Matrix protein 2,External core antigen / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Hepatitis B virus / Strain: isolate France/Tiollais/1979 Hepatitis B virus / Strain: isolate France/Tiollais/1979 |
| Molecular weight | Theoretical: 30.959662 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MDPYKEFGAT VELLSFLPSD FFPSVRDLLD TASALYREAL ESPEHCSPHH TALRQAILCW GELMTLATWV GGNLEDGSGT SGSSGSGSG GSGSGGGGEL SLLTEVETPI RNEWGSRSND SSDDLSLLTE VETPIRNEWG SRSNDSSDDL SLLTEVETPI R NEWGSRSN ...String: MDPYKEFGAT VELLSFLPSD FFPSVRDLLD TASALYREAL ESPEHCSPHH TALRQAILCW GELMTLATWV GGNLEDGSGT SGSSGSGSG GSGSGGGGEL SLLTEVETPI RNEWGSRSND SSDDLSLLTE VETPIRNEWG SRSNDSSDDL SLLTEVETPI R NEWGSRSN DSSDDLSLLT EVETPIRNEW GSRSNDSSDD IGTSGSSGSG SGGSGSGGGG GPSRDLVVSY VNTNMGLKFR QL LWFHISC LTFGRETVIE YLVSFGVWIR TPPAYRPPNA PILSTLPETT VVC |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.00026000000000000003 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 0.05 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.2 µm Bright-field microscopy / Cs: 0.05 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.2 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 60.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION |
|---|---|
| Final angle assignment | Type: OTHER / Software - Name: cryoSPARC |
| Final reconstruction | Applied symmetry - Point group: I (icosahedral ) / Resolution.type: BY AUTHOR / Resolution: 4.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 4757 ) / Resolution.type: BY AUTHOR / Resolution: 4.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 4757 |
 Movie
Movie Controller
Controller






 Z
Z Y
Y X
X