[English] 日本語
 Yorodumi
Yorodumi- EMDB-1585: Electron tomography of isometrically contracting insect flight mu... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1585 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Electron tomography of isometrically contracting insect flight muscle quick frozen after a rapid stretch transient | |||||||||
 Map data Map data | This is a global average of aligned subvolumes. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  insect / insect /  muscle / muscle /  myosin / myosin /  troponin / troponin /  tropomyosin / tropomyosin /  actin / light chains / actin / light chains /  thin filament / thin filament /  thick filament / thick filament /  electron microscopy / electron microscopy /  image processing / image processing /  isometric contraction / isometric contraction /  freezing / freeze substitution / freezing / freeze substitution /  microtomy / microtomy /  multivariate data analysis multivariate data analysis | |||||||||
| Function / homology |  Function and homology information Function and homology information troponin C binding / contractile muscle fiber / troponin C binding / contractile muscle fiber /  troponin complex / Striated Muscle Contraction / troponin complex / Striated Muscle Contraction /  regulation of muscle contraction / myosin filament / regulation of muscle contraction / myosin filament /  myosin II complex / myosin II complex /  myosin complex / sarcomere organization / cytoskeletal motor activator activity ... myosin complex / sarcomere organization / cytoskeletal motor activator activity ... troponin C binding / contractile muscle fiber / troponin C binding / contractile muscle fiber /  troponin complex / Striated Muscle Contraction / troponin complex / Striated Muscle Contraction /  regulation of muscle contraction / myosin filament / regulation of muscle contraction / myosin filament /  myosin II complex / myosin II complex /  myosin complex / sarcomere organization / cytoskeletal motor activator activity / microfilament motor activity / myosin complex / sarcomere organization / cytoskeletal motor activator activity / microfilament motor activity /  tropomyosin binding / tropomyosin binding /  myofibril / myofibril /  myosin heavy chain binding / mesenchyme migration / myosin heavy chain binding / mesenchyme migration /  troponin I binding / actin filament bundle / filamentous actin / actin filament bundle assembly / skeletal muscle thin filament assembly / cytoskeletal motor activity / striated muscle thin filament / skeletal muscle myofibril / skeletal muscle contraction / actin monomer binding / skeletal muscle fiber development / skeletal muscle tissue development / troponin I binding / actin filament bundle / filamentous actin / actin filament bundle assembly / skeletal muscle thin filament assembly / cytoskeletal motor activity / striated muscle thin filament / skeletal muscle myofibril / skeletal muscle contraction / actin monomer binding / skeletal muscle fiber development / skeletal muscle tissue development /  stress fiber / stress fiber /  titin binding / cardiac muscle contraction / actin filament polymerization / titin binding / cardiac muscle contraction / actin filament polymerization /  filopodium / filopodium /  muscle contraction / muscle contraction /  actin filament / actin filament /  Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / calcium-dependent protein binding / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / calcium-dependent protein binding /  actin filament binding / actin filament binding /  actin cytoskeleton / actin cytoskeleton /  lamellipodium / lamellipodium /  cell body / cell body /  actin binding / actin binding /  calmodulin binding / calmodulin binding /  hydrolase activity / protein heterodimerization activity / protein domain specific binding / hydrolase activity / protein heterodimerization activity / protein domain specific binding /  calcium ion binding / positive regulation of gene expression / magnesium ion binding / protein homodimerization activity / calcium ion binding / positive regulation of gene expression / magnesium ion binding / protein homodimerization activity /  ATP binding / identical protein binding / ATP binding / identical protein binding /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Lethocerus indicus (insect) Lethocerus indicus (insect) | |||||||||
| Method | subtomogram averaging /  cryo EM / cryo EM /  negative staining negative staining | |||||||||
 Authors Authors | Wu S / Liu J / Reedy MC / Tregear RT / Winkler H / Franzini-Armstrong C / Sasaki H / Lucaveche C / Goldman YE / Reedy MK / Taylor KA | |||||||||
 Citation Citation | Journal: J Struct Biol / Year: 2009 Title: Methods for identifying and averaging variable molecular conformations in tomograms of actively contracting insect flight muscle. Authors: Shenping Wu / Jun Liu / Mary C Reedy / Hanspeter Winkler / Michael K Reedy / Kenneth A Taylor /  Abstract: During active muscle contraction, tension is generated through many simultaneous, independent interactions between the molecular motor myosin and the actin filaments. The ensemble of myosin motors ...During active muscle contraction, tension is generated through many simultaneous, independent interactions between the molecular motor myosin and the actin filaments. The ensemble of myosin motors displays heterogeneous conformations reflecting different mechanochemical steps of the ATPase pathway. We used electron tomography of actively contracting insect flight muscle fast-frozen, freeze substituted, Araldite embedded, thin-sectioned and stained, to obtain 3D snapshots of the multiplicity of actin-attached myosin structures. We describe procedures for alignment of the repeating lattice of sub-volumes (38.7 nm cross-bridge repeats bounded by troponin) and multivariate data analysis to identify self-similar repeats for computing class averages. Improvements in alignment and classification of repeat sub-volumes reveals (for the first time in active muscle images) the helix of actin subunits in the thin filament and the troponin density with sufficient clarity that a quasiatomic model of the thin filament can be built into the class averages independent of the myosin cross-bridges. We show how quasiatomic model building can identify both strong and weak myosin attachments to actin. We evaluate the accuracy of image classification to enumerate the different types of actin-myosin attachments. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1585.map.gz emd_1585.map.gz | 1.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1585-v30.xml emd-1585-v30.xml emd-1585.xml emd-1585.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-1585.png EMD-1585.png | 86.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1585 http://ftp.pdbj.org/pub/emdb/structures/EMD-1585 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1585 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1585 | HTTPS FTP |
-Related structure data
| Related structure data |  2w4gM  2w4hMC  2w4uMC  2w4vMC  2w4wMC  1584C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1585.map.gz / Format: CCP4 / Size: 1.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1585.map.gz / Format: CCP4 / Size: 1.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a global average of aligned subvolumes. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 6.9 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
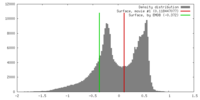
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Isometrically contracting asynchronous insect flight muscle upon ...
| Entire | Name: Isometrically contracting asynchronous insect flight muscle upon a quick stretch |
|---|---|
| Components |
|
-Supramolecule #1000: Isometrically contracting asynchronous insect flight muscle upon ...
| Supramolecule | Name: Isometrically contracting asynchronous insect flight muscle upon a quick stretch type: sample / ID: 1000 Details: This specimen is obtained from a quick frozen, isometrically contracting asynchronous insect flight muscle that has been freeze substituted, plastic embedded, and thin sectioned. The fiber ...Details: This specimen is obtained from a quick frozen, isometrically contracting asynchronous insect flight muscle that has been freeze substituted, plastic embedded, and thin sectioned. The fiber was stretched 6 nm per half-sarcomere in 2 ms and length was held constant after the length step. The freezing impact occurred 6-7 ms later. Oligomeric state: tissue / Number unique components: 8 |
|---|
-Supramolecule #1: myofibril
| Supramolecule | Name: myofibril / type: organelle_or_cellular_component / ID: 1 / Name.synonym: muscle Details: The sample was enblock stained using uranyl acetate and tannic acid, and post stained with lead citrate and potassium permanganate Oligomeric state: muscle fibril / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:   Lethocerus indicus (insect) / synonym: Water bug / Tissue: asynchronous dorsal longitudinal flight muscle / Organelle: myofibril / Location in cell: myoplasm Lethocerus indicus (insect) / synonym: Water bug / Tissue: asynchronous dorsal longitudinal flight muscle / Organelle: myofibril / Location in cell: myoplasm |
-Experimental details
-Structure determination
| Method |  negative staining, negative staining,  cryo EM cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
- Sample preparation
Sample preparation
| Buffer | Details: 20 mM MOPS buffer, 5 mM NaN3, and MgCl2, ATP, CaCl2, and EGTA in varying millimolar concentrations |
|---|---|
| Staining | Type: NEGATIVE Details: Freeze slammed fibers were freeze-substituted in acetone using a tannic aciduranyl acetate sequence, and ultimately embedded in Araldite-506 for thin-section electron microscopy. Ultrathin ...Details: Freeze slammed fibers were freeze-substituted in acetone using a tannic aciduranyl acetate sequence, and ultimately embedded in Araldite-506 for thin-section electron microscopy. Ultrathin (25-30 nm) longitudinal sections were stained by permanganate-lead. |
| Vitrification | Cryogen name: HELIUM / Chamber temperature: 4.5 K / Instrument: HOMEMADE PLUNGER Details: Vitrification instrument: modified Heuser Cryopress freezing head Timed resolved state: Frozen 6-7 msec after application of the length transient Method: smash against a liquid helium cooled Au-coated Cu mirror |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM300FEG/T |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.0 mm Bright-field microscopy / Cs: 2.0 mm |
| Sample stage | Specimen holder: side entry, eucentric / Specimen holder model: OTHER / Tilt series - Axis1 - Min angle: -72 ° / Tilt series - Axis1 - Max angle: 72 ° |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F224 (2k x 2k) Details: Images recorded on a TVIPS Tem-Cam F224 2k x 2k CCD camera Bits/pixel: 16 |
- Image processing
Image processing
| Final reconstruction | Algorithm: OTHER / Resolution method: FSC 0.5 CUT-OFF / Software - Name: PROTOMO, IMOD Details: Two tilt series at 90 degrees to one another were first independently aligned using marker-free alignment and area matching. The two resulting tomograms were then merged by patch correlation ...Details: Two tilt series at 90 degrees to one another were first independently aligned using marker-free alignment and area matching. The two resulting tomograms were then merged by patch correlation and volume warp using IMOD. Tomogram computed using weighted back projection. |
|---|---|
| Details | The fiber was stretched 6 nm per half-sarcomere in 2 ms and length was held constant after the length step. The freezing impact occurred 6-7 ms later. Average number of tilts used in the 3D reconstructions: 70. |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)