+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | p97 with I3 substrate in channel | ||||||||||||
 Map data Map data | p97 map plus I3 substrate in channel | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | AAA+ ATPase / p97 / VCP / Cdc48 / unfoldase /  protein phosphatase-1 / protein maturation / protein phosphatase-1 / protein maturation /  CHAPERONE CHAPERONE | ||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||||||||
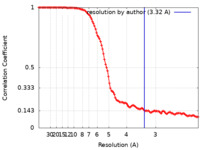
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.32 Å cryo EM / Resolution: 3.32 Å | ||||||||||||
 Authors Authors | Saibil HR / van den Boom J / Marini G / Meyer H | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: EMBO J / Year: 2023 Journal: EMBO J / Year: 2023Title: Structural basis of ubiquitin-independent PP1 complex disassembly by p97. Authors: Johannes van den Boom / Guendalina Marini / Hemmo Meyer / Helen R Saibil /   Abstract: The AAA+-ATPase p97 (also called VCP or Cdc48) unfolds proteins and disassembles protein complexes in numerous cellular processes, but how substrate complexes are loaded onto p97 and disassembled is ...The AAA+-ATPase p97 (also called VCP or Cdc48) unfolds proteins and disassembles protein complexes in numerous cellular processes, but how substrate complexes are loaded onto p97 and disassembled is unclear. Here, we present cryo-EM structures of p97 in the process of disassembling a protein phosphatase-1 (PP1) complex by extracting an inhibitory subunit from PP1. We show that PP1 and its partners SDS22 and inhibitor-3 (I3) are loaded tightly onto p97, surprisingly via a direct contact of SDS22 with the p97 N-domain. Loading is assisted by the p37 adapter that bridges two adjacent p97 N-domains underneath the substrate complex. A stretch of I3 is threaded into the central channel of the spiral-shaped p97 hexamer, while other elements of I3 are still attached to PP1. Thus, our data show how p97 arranges a protein complex between the p97 N-domain and central channel, suggesting a hold-and-extract mechanism for p97-mediated disassembly. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15774.map.gz emd_15774.map.gz | 98.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15774-v30.xml emd-15774-v30.xml emd-15774.xml emd-15774.xml | 22.2 KB 22.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15774_fsc.xml emd_15774_fsc.xml | 14.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_15774.png emd_15774.png | 158.8 KB | ||
| Others |  emd_15774_half_map_1.map.gz emd_15774_half_map_1.map.gz emd_15774_half_map_2.map.gz emd_15774_half_map_2.map.gz | 98.4 MB 98.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15774 http://ftp.pdbj.org/pub/emdb/structures/EMD-15774 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15774 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15774 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15774.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15774.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | p97 map plus I3 substrate in channel | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map 2 unmasked
| File | emd_15774_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 unmasked | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1 unmasked
| File | emd_15774_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 unmasked | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : p97 - substrate complex, map of p97 with part of I3 in channel only
| Entire | Name: p97 - substrate complex, map of p97 with part of I3 in channel only |
|---|---|
| Components |
|
-Supramolecule #1: p97 - substrate complex, map of p97 with part of I3 in channel only
| Supramolecule | Name: p97 - substrate complex, map of p97 with part of I3 in channel only type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: p97, SDS22, PP1 gamma and I3
| Supramolecule | Name: p97, SDS22, PP1 gamma and I3 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: p37
| Supramolecule | Name: p37 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #5 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: p97
| Macromolecule | Name: p97 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number:  vesicle-fusing ATPase vesicle-fusing ATPase |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASGADSKGD DLSTAILKQK NRPNRLIVDE AINEDNSVVS LSQPKMDELQ LFRGDTVLLK GKKRREAVCI VLSDDTCSDE KIRMNRVVRN NLRVRLGDVI SIQPCPDVKY GKRIHVLPID DTVEGITGNL FEVYLKPYFL EAYRPIRKGD IFLVRGGMRA VEFKVVETDP ...String: MASGADSKGD DLSTAILKQK NRPNRLIVDE AINEDNSVVS LSQPKMDELQ LFRGDTVLLK GKKRREAVCI VLSDDTCSDE KIRMNRVVRN NLRVRLGDVI SIQPCPDVKY GKRIHVLPID DTVEGITGNL FEVYLKPYFL EAYRPIRKGD IFLVRGGMRA VEFKVVETDP SPYCIVAPDT VIHCEGEPIK REDEEESLNE VGYDDIGGCR KQLAQIKEMV ELPLRHPALF KAIGVKPPRG ILLYGPPGTG KTLIARAVAN ETGAFFFLIN GPEIMSKLAG ESESNLRKAF EEAEKNAPAI IFIDELDAIA PKREKTHGEV ERRIVSQLLT LMDGLKQRAH VIVMAATNRP NSIDPALRRF GRFDREVDIG IPDATGRLEI LQIHTKNMKL ADDVDLEQVA NETHGHVGAD LAALCSEAAL QAIRKKMDLI DLEDETIDAE VMNSLAVTMD DFRWALSQSN PSALRETVVE VPQVTWEDIG GLEDVKRELQ ELVQYPVEHP DKFLKFGMTP SKGVLFYGPP GCGKTLLAKA IANECQANFI SIKGPELLTM WFGESEANVR EIFDKARQAA PCVLFFDELD SIAKARGGNI GDGGGAADRV INQILTEMDG MSTKKNVFII GATNRPDIID PAILRPGRLD QLIYIPLPDE KSRVAILKAN LRKSPVAKDV DLEFLAKMTN GFSGADLTEI CQRACKLAIR ESIESEIRRE RERQTNPSAM EVEEDDPVPE IRRDHFEEAM RFARRSVSDN DIRKYEMFAQ TLQQSRGFGS FRFPSGNQGG AGPSQGSGGG TGGSVYTEDN DDDLYG |
-Macromolecule #2: SDS22
| Macromolecule | Name: SDS22 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAAERGAGQQ QSQEMMEVDR RVESEESGDE EGKKHSSGIV ADLSEQSLKD GEERGEEDPE EEHELPVDME TINLDRDAED VDLNHYRIGK IEGFEVLKKV KTLCLRQNLI KCIENLEELQ SLRELDLYDN QIKKIENLEA LTELEILDIS FNLLRNIEGV DKLTRLKKLF ...String: MAAERGAGQQ QSQEMMEVDR RVESEESGDE EGKKHSSGIV ADLSEQSLKD GEERGEEDPE EEHELPVDME TINLDRDAED VDLNHYRIGK IEGFEVLKKV KTLCLRQNLI KCIENLEELQ SLRELDLYDN QIKKIENLEA LTELEILDIS FNLLRNIEGV DKLTRLKKLF LVNNKISKIE NLSNLHQLQM LELGSNRIRA IENIDTLTNL ESLFLGKNKI TKLQNLDALT NLTVLSMQSN RLTKIEGLQN LVNLRELYLS HNGIEVIEGL ENNNKLTMLD IASNRIKKIE NISHLTELQE FWMNDNLLES WSDLDELKGA RSLETVYLER NPLQKDPQYR RKVMLALPSV RQIDATFVRF |
-Macromolecule #3: PP1 gamma
| Macromolecule | Name: PP1 gamma / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO / EC number: protein-serine/threonine phosphatase |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MADLDKLNID SIIQRLLEVR GSKPGKNVQL QENEIRGLCL KSREIFLSQP ILLELEAPLK ICGDIHGQYY DLLRLFEYGG FPPESNYLFL GDYVDRGKQS LETICLLLAY KIKYPENFFL LRGNHECASI NRIYGFYDEC KRRYNIKLWK TFTDCFNCLP IAAIVDEKIF ...String: MADLDKLNID SIIQRLLEVR GSKPGKNVQL QENEIRGLCL KSREIFLSQP ILLELEAPLK ICGDIHGQYY DLLRLFEYGG FPPESNYLFL GDYVDRGKQS LETICLLLAY KIKYPENFFL LRGNHECASI NRIYGFYDEC KRRYNIKLWK TFTDCFNCLP IAAIVDEKIF CCHGGLSPDL QSMEQIRRIM RPTDVPDQGL LCDLLWSDPD KDVLGWGEND RGVSFTFGAE VVAKFLHKHD LDLICRAHQV VEDGYEFFAK RQLVTLFSAP NYCGEFDNAG AMMSVDETLM CSFQILKPAE KKKPNATRPV TPPRGMITKQ AKK |
-Macromolecule #4: I3
| Macromolecule | Name: I3 / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSYYHHHHHH DYDIPTTENL YFQGAMGSMA EAGAGLSETV TETTVTVTTE PENRSLTIKL RKRKPEKKVE WTSDTVDNEH MGRRSSKCCC IYEKPRAFGE SSTESDEEEE EGCGHTHCVR GHRKGRRRAT LGPTPTTPPQ PPDPSQPPPG PMQH |
-Macromolecule #5: p37
| Macromolecule | Name: p37 / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: GPLGSMAEGG RAEPEEQERG SSRPRPPSAR DLQLALAELY EDEMKCKSSK PDRSTPATCR SPRTPPHRLY SGDHKYDGLH IVQPPTGKIV NELFKEAREH GAVPLNEATR SSREDKTKSF TGGGYRLGNS FYKRSEYIYG ENQLQDVQVL LKLWRNGFSL DDGELRPYSD ...String: GPLGSMAEGG RAEPEEQERG SSRPRPPSAR DLQLALAELY EDEMKCKSSK PDRSTPATCR SPRTPPHRLY SGDHKYDGLH IVQPPTGKIV NELFKEAREH GAVPLNEATR SSREDKTKSF TGGGYRLGNS FYKRSEYIYG ENQLQDVQVL LKLWRNGFSL DDGELRPYSD PTNAQFLESV KRGETPLELQ RLVHGAQVNL DMEDHQDQEY IKPRLRFKAF SGEGQKLGSL TPEIVSTPSS PEEEDKSILN AAVLIDDSMP TTKIQIRLAD GSRLVQRFNS THRILDVRDF IVRSRPEFAT TDFILVTSFP SKELTDETVT LQEADILNTV ILQQLK |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Details: 50 mM HEPES pH 7.5, 150 mM KCl, 5 mM MgCl2, 2% glycerol, 1 mM DTT, 0.0005% NP40 and 2 mM ADP-BeFx |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.3000000000000003 µm / Nominal defocus min: 1.8 µm / Nominal magnification: 81000 Bright-field microscopy / Nominal defocus max: 3.3000000000000003 µm / Nominal defocus min: 1.8 µm / Nominal magnification: 81000 |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average exposure time: 3.0 sec. / Average electron dose: 48.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)