[English] 日本語
 Yorodumi
Yorodumi- EMDB-15690: Cryo-EM structure of the Tb ADAT2/3 deaminase in complex with tRNA -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Tb ADAT2/3 deaminase in complex with tRNA | |||||||||
 Map data Map data | Final non b factor sharpened cryosparc map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  ADAT / ADAT /  inosine / tRNA modification / inosine / tRNA modification /  deaminase / cryo-EM structure / deaminase / cryo-EM structure /  Trypanosoma brucei / Trypanosoma brucei /  RNA BINDING PROTEIN RNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationtRNA wobble adenosine to inosine editing / tRNA-specific adenosine-34 deaminase activity /  Hydrolases; Acting on carbon-nitrogen bonds, other than peptide bonds; In cyclic amidines / Hydrolases; Acting on carbon-nitrogen bonds, other than peptide bonds; In cyclic amidines /  hydrolase activity / zinc ion binding / hydrolase activity / zinc ion binding /  nucleoplasm / nucleoplasm /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Trypanosoma brucei brucei (eukaryote) / Trypanosoma brucei brucei (eukaryote) /   Trypanosoma brucei (eukaryote) Trypanosoma brucei (eukaryote) | |||||||||
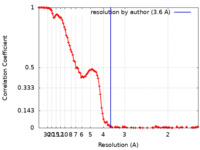
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.6 Å cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Dolce LG / Tengo L / Weis F / Kowalinski E | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural basis for sequence-independent substrate selection by eukaryotic wobble base tRNA deaminase ADAT2/3. Authors: Luciano G Dolce / Aubree A Zimmer / Laura Tengo / Félix Weis / Mary Anne T Rubio / Juan D Alfonzo / Eva Kowalinski /    Abstract: The essential deamination of adenosine A to inosine at the wobble base is the individual tRNA modification with the greatest effects on mRNA decoding, empowering a single tRNA to translate three ...The essential deamination of adenosine A to inosine at the wobble base is the individual tRNA modification with the greatest effects on mRNA decoding, empowering a single tRNA to translate three different codons. To date, many aspects of how eukaryotic deaminases specifically select their multiple substrates remain unclear. Here, using cryo-EM, we present the structure of a eukaryotic ADAT2/3 deaminase bound to a full-length tRNA, revealing that the enzyme distorts the anticodon loop, but in contrast to the bacterial enzymes, selects its substrate via sequence-independent contacts of eukaryote-acquired flexible or intrinsically unfolded motifs distal from the conserved catalytic core. A gating mechanism for substrate entry to the active site is identified. Our multi-step tRNA recognition model yields insights into how RNA editing by A deamination evolved, shaped the genetic code, and directly impacts the eukaryotic proteome. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15690.map.gz emd_15690.map.gz | 50.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15690-v30.xml emd-15690-v30.xml emd-15690.xml emd-15690.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15690_fsc.xml emd_15690_fsc.xml | 13.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_15690.png emd_15690.png | 51.3 KB | ||
| Filedesc metadata |  emd-15690.cif.gz emd-15690.cif.gz | 6.2 KB | ||
| Others |  emd_15690_additional_1.map.gz emd_15690_additional_1.map.gz emd_15690_half_map_1.map.gz emd_15690_half_map_1.map.gz emd_15690_half_map_2.map.gz emd_15690_half_map_2.map.gz | 90.3 MB 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15690 http://ftp.pdbj.org/pub/emdb/structures/EMD-15690 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15690 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15690 | HTTPS FTP |
-Related structure data
| Related structure data |  8aw3MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15690.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15690.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final non b factor sharpened cryosparc map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.81 Å | ||||||||||||||||||||||||||||||||||||
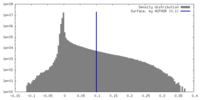
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: deepEMhancer post processed map
| File | emd_15690_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | deepEMhancer post processed map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A
| File | emd_15690_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_15690_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ADAT2/3 in complex with tRNA
| Entire | Name: ADAT2/3 in complex with tRNA |
|---|---|
| Components |
|
-Supramolecule #1: ADAT2/3 in complex with tRNA
| Supramolecule | Name: ADAT2/3 in complex with tRNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Trypanosoma brucei brucei (eukaryote) Trypanosoma brucei brucei (eukaryote) |
| Molecular weight | Theoretical: 90 KDa |
-Macromolecule #1: RNA (75-MER)
| Macromolecule | Name: RNA (75-MER) / type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:   Trypanosoma brucei (eukaryote) Trypanosoma brucei (eukaryote) |
| Molecular weight | Theoretical: 24.132311 KDa |
| Sequence | String: GGCCGCUUAG CACAGUGGCA GUGCACCACU CUCGUAAAGU GGGGGUCGCG AGUUCGAUUC UCGCAGUGGC CUCCA GENBANK:  GENBANK: AC091702.3 GENBANK: AC091702.3 |
-Macromolecule #2: Deaminase, putative
| Macromolecule | Name: Deaminase, putative / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO EC number:  Hydrolases; Acting on carbon-nitrogen bonds, other than peptide bonds; In cyclic amidines Hydrolases; Acting on carbon-nitrogen bonds, other than peptide bonds; In cyclic amidines |
|---|---|
| Source (natural) | Organism:   Trypanosoma brucei brucei (eukaryote) / Strain: 927/4 GUTat10.1 Trypanosoma brucei brucei (eukaryote) / Strain: 927/4 GUTat10.1 |
| Molecular weight | Theoretical: 23.547535 KDa |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MVQDTGKDTN LKGTAEANES VVYCDVFMQA ALKEATCALE EGEVPVGCVL VKADSSTAAQ AQAGDDLALQ KLIVARGRNA TNRKGHGLA HAEFVAVEEL LRQATAGTSE NIGGGGNSGA VSQDLADYVL YVVVEPCIMC AAMLLYNRVR KVYFGCTNPR F GGNGTVLS ...String: MVQDTGKDTN LKGTAEANES VVYCDVFMQA ALKEATCALE EGEVPVGCVL VKADSSTAAQ AQAGDDLALQ KLIVARGRNA TNRKGHGLA HAEFVAVEEL LRQATAGTSE NIGGGGNSGA VSQDLADYVL YVVVEPCIMC AAMLLYNRVR KVYFGCTNPR F GGNGTVLS VHNSYKGCSG EDAALIGYES CGGYRAEEAV VLLQQFYRRE NTNAPLGKRK RKD UniProtKB:  Deaminase, putative Deaminase, putative |
-Macromolecule #3: Deaminase, putative
| Macromolecule | Name: Deaminase, putative / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Trypanosoma brucei brucei (eukaryote) / Strain: 927/4 GUTat10.1 Trypanosoma brucei brucei (eukaryote) / Strain: 927/4 GUTat10.1 |
| Molecular weight | Theoretical: 40.600254 KDa |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GPDSMEEVVV PEEPPKLVSA LATYVQQERL CTMFLSIANK LLPLKPHACH LKRIRRSSAT RVATAPMDGF AGGVICDKRD SSVATSTIS DGCERNSAAL GTPAAEKSHV LELLLSVGGP VDSSALKELE SAADTTVAVH RVWVPDRAPR SSAEEWTKWC Q IWPFATPK ...String: GPDSMEEVVV PEEPPKLVSA LATYVQQERL CTMFLSIANK LLPLKPHACH LKRIRRSSAT RVATAPMDGF AGGVICDKRD SSVATSTIS DGCERNSAAL GTPAAEKSHV LELLLSVGGP VDSSALKELE SAADTTVAVH RVWVPDRAPR SSAEEWTKWC Q IWPFATPK PRVPTQLSEC EVGSIQRIFR TVVMPLAKRL RTDETLGIAA VLVDPSDGYR VLVSSGEEHA LKRGNSAACL GY VSNSGCR KSNRVVLDHP VTFVLKEVTR KQCKDREVEG DASYLANGMD MFVSHEPCVM CSMALVHSRV RRVFYCFPNP VHG GLGSTV SIHAIQELNH HFRVFRCDSR WLSDPEGVSS DHDNPYWEDL TVP UniProtKB:  Deaminase, putative Deaminase, putative |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.07 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 55.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)