[English] 日本語
 Yorodumi
Yorodumi- EMDB-1542: Three-dimensional structure of Pyrococcus furiosus A1Ao ATP synthase -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1542 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
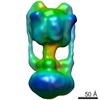
| Title | Three-dimensional structure of Pyrococcus furiosus A1Ao ATP synthase | |||||||||
 Map data Map data | volume of Pyrococcus furiosus A-type ATP synthase | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  ATP synthase / A1Ao ATP synthase ATP synthase / A1Ao ATP synthase | |||||||||
| Biological species |    Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 23.0 Å negative staining / Resolution: 23.0 Å | |||||||||
 Authors Authors | Vonck J / Pisa KY / Morgner N / Brutschy B / Mueller V | |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2009 Journal: J Biol Chem / Year: 2009Title: Three-dimensional structure of A1A0 ATP synthase from the hyperthermophilic archaeon Pyrococcus furiosus by electron microscopy. Authors: Janet Vonck / Kim Y Pisa / Nina Morgner / Bernhard Brutschy / Volker Müller /  Abstract: The archaeal ATP synthase is a multisubunit complex that consists of a catalytic A(1) part and a transmembrane, ion translocation domain A(0). The A(1)A(0) complex from the hyperthermophile ...The archaeal ATP synthase is a multisubunit complex that consists of a catalytic A(1) part and a transmembrane, ion translocation domain A(0). The A(1)A(0) complex from the hyperthermophile Pyrococcus furiosus was isolated. Mass analysis of the complex by laser-induced liquid bead ion desorption (LILBID) indicated a size of 730 +/- 10 kDa. A three-dimensional map was generated by electron microscopy from negatively stained images. The map at a resolution of 2.3 nm shows the A(1) and A(0) domain, connected by a central stalk and two peripheral stalks, one of which is connected to A(0), and both connected to A(1) via prominent knobs. X-ray structures of subunits from related proteins were fitted to the map. On the basis of the fitting and the LILBID analysis, a structural model is presented with the stoichiometry A(3)B(3)CDE(2)FH(2)ac(10). | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1542.map.gz emd_1542.map.gz | 516.8 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1542-v30.xml emd-1542-v30.xml emd-1542.xml emd-1542.xml | 9.6 KB 9.6 KB | Display Display |  EMDB header EMDB header |
| Images |  1542.gif 1542.gif | 19.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1542 http://ftp.pdbj.org/pub/emdb/structures/EMD-1542 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1542 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1542 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1542.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1542.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | volume of Pyrococcus furiosus A-type ATP synthase | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.77 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : A1Ao ATP synthase from Pyrococcus furiosus
| Entire | Name: A1Ao ATP synthase from Pyrococcus furiosus |
|---|---|
| Components |
|
-Supramolecule #1000: A1Ao ATP synthase from Pyrococcus furiosus
| Supramolecule | Name: A1Ao ATP synthase from Pyrococcus furiosus / type: sample / ID: 1000 / Oligomeric state: nine different subunits / Number unique components: 9 |
|---|---|
| Molecular weight | Experimental: 738 KDa / Theoretical: 738 KDa / Method: LILBID mass spectroscopy |
-Macromolecule #1: A1Ao ATP synthase
| Macromolecule | Name: A1Ao ATP synthase / type: protein_or_peptide / ID: 1 / Name.synonym: ATP synthase / Oligomeric state: heteromultimer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:    Pyrococcus furiosus (archaea) / Cell: archaeon / Location in cell: plasma membrane Pyrococcus furiosus (archaea) / Cell: archaeon / Location in cell: plasma membrane |
| Molecular weight | Experimental: 738 KDa / Theoretical: 738 KDa |
-Macromolecule #2: A1Ao ATP synthase
| Macromolecule | Name: A1Ao ATP synthase / type: protein_or_peptide / ID: 2 / Name.synonym: ATP synthase / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:    Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) |
| Molecular weight | Experimental: 738 KDa / Theoretical: 738 KDa |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 50 mM this/HCl, 5 mM MgCl2, 10% glycerol, 50 mM KCl, 0.1% Triton-X100, 0.1 mM PMSF |
| Staining | Type: NEGATIVE Details: a drop of 1% (w/v) uranyl acetate was added to a carbon-coated grid with absorbed protein and blotted after 30 s. |
| Grid | Details: 400 mesh copper grid |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM120T |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 44000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2 mm / Nominal defocus max: 0.4 µm / Nominal defocus min: 0.4 µm / Nominal magnification: 45000 Bright-field microscopy / Cs: 2 mm / Nominal defocus max: 0.4 µm / Nominal defocus min: 0.4 µm / Nominal magnification: 45000 |
| Sample stage | Specimen holder: eucentric / Specimen holder model: OTHER |
| Alignment procedure | Legacy - Astigmatism: objective lens astigmatism was corrected at 160,000 times magnification |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Number real images: 51 / Average electron dose: 20 e/Å2 / Bits/pixel: 8 |
- Image processing
Image processing
| Final two d classification | Number classes: 392 |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 23.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN / Number images used: 12912 |
 Movie
Movie Controller
Controller