[English] 日本語
 Yorodumi
Yorodumi- EMDB-14845: cryo-EM structure of CGT ABC transporter in presence of CBG substrate -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM structure of CGT ABC transporter in presence of CBG substrate | ||||||||||||
 Map data Map data | sugar | ||||||||||||
 Sample Sample |
| ||||||||||||
| Function / homology |  Function and homology information Function and homology informationABC-type beta-glucan transporter / ABC-type beta-glucan transporter activity /  : / : /  ATP hydrolysis activity / ATP hydrolysis activity /  ATP binding / ATP binding /  plasma membrane plasma membraneSimilarity search - Function | ||||||||||||
| Biological species |   Brucella abortus 2308 (bacteria) Brucella abortus 2308 (bacteria) | ||||||||||||
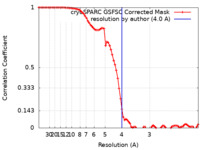
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.0 Å cryo EM / Resolution: 4.0 Å | ||||||||||||
 Authors Authors | Jaroslaw S / Dong CN / Frank L / Na W / Renato Z / Seunho J / Henning S / Christoph D | ||||||||||||
| Funding support |  Switzerland, 3 items Switzerland, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Mechanism of cyclic β-glucan export by ABC transporter Cgt of Brucella. Authors: Jaroslaw Sedzicki / Dongchun Ni / Frank Lehmann / Na Wu / Renato Zenobi / Seunho Jung / Henning Stahlberg / Christoph Dehio /   Abstract: Polysaccharides play critical roles in bacteria, including the formation of protective capsules and biofilms and establishing specific host cell interactions. Their transport across membranes is ...Polysaccharides play critical roles in bacteria, including the formation of protective capsules and biofilms and establishing specific host cell interactions. Their transport across membranes is often mediated by ATP-binding cassette (ABC) transporters, which utilize ATP to translocate diverse molecules. Cyclic β-glucans (CβGs) are critical for host interaction of the Rhizobiales, including the zoonotic pathogen Brucella. CβGs are exported into the periplasmic space by the cyclic glucan transporter (Cgt). The interaction of an ABC transporter with a polysaccharide substrate has not been visualized so far. Here we use single-particle cryoelectron microscopy to elucidate the structures of Cgt from Brucella abortus in four conformational states. The substrate-bound structure reveals an unusual binding pocket at the height of the cytoplasmic leaflet, whereas ADP-vanadate models hint at an alternative mechanism of substrate release. Our work provides insights into the translocation of large, heterogeneous substrates and sheds light on protein-polysaccharide interactions in general. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14845.map.gz emd_14845.map.gz | 49.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14845-v30.xml emd-14845-v30.xml emd-14845.xml emd-14845.xml | 24.5 KB 24.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14845_fsc.xml emd_14845_fsc.xml | 7.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_14845.png emd_14845.png | 70.9 KB | ||
| Others |  emd_14845_additional_1.map.gz emd_14845_additional_1.map.gz emd_14845_additional_2.map.gz emd_14845_additional_2.map.gz emd_14845_additional_3.map.gz emd_14845_additional_3.map.gz emd_14845_half_map_1.map.gz emd_14845_half_map_1.map.gz emd_14845_half_map_2.map.gz emd_14845_half_map_2.map.gz | 49.7 MB 49.7 MB 49.6 MB 49 MB 49 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14845 http://ftp.pdbj.org/pub/emdb/structures/EMD-14845 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14845 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14845 | HTTPS FTP |
-Related structure data
| Related structure data |  7zoaMC  7znuC  7zo8C  7zo9C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14845.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14845.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sugar | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.025 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: sugar
| File | emd_14845_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sugar | ||||||||||||
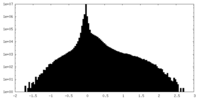
| Projections & Slices |
| ||||||||||||
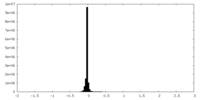
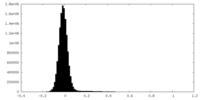
| Density Histograms |
-Additional map: sugar
| File | emd_14845_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sugar | ||||||||||||
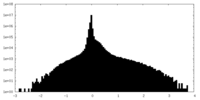
| Projections & Slices |
| ||||||||||||
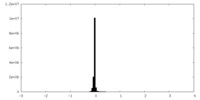
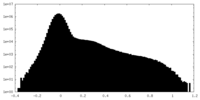
| Density Histograms |
-Additional map: sugar
| File | emd_14845_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sugar | ||||||||||||
| Projections & Slices |
| ||||||||||||
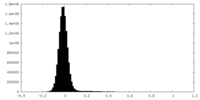
| Density Histograms |
-Half map: sugar
| File | emd_14845_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sugar | ||||||||||||
| Projections & Slices |
| ||||||||||||
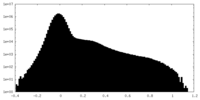
| Density Histograms |
-Half map: sugar
| File | emd_14845_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sugar | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of CGT ABC transporter with the substrate Cyclic beta glucan
| Entire | Name: Complex of CGT ABC transporter with the substrate Cyclic beta glucan |
|---|---|
| Components |
|
-Supramolecule #1: Complex of CGT ABC transporter with the substrate Cyclic beta glucan
| Supramolecule | Name: Complex of CGT ABC transporter with the substrate Cyclic beta glucan type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Brucella abortus 2308 (bacteria) Brucella abortus 2308 (bacteria) |
| Molecular weight | Theoretical: 125 KDa |
-Macromolecule #1: Beta-(1-->2)glucan export ATP-binding/permease protein NdvA
| Macromolecule | Name: Beta-(1-->2)glucan export ATP-binding/permease protein NdvA type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: ABC-type beta-glucan transporter |
|---|---|
| Source (natural) | Organism:   Brucella abortus 2308 (bacteria) / Strain: 2308 Brucella abortus 2308 (bacteria) / Strain: 2308 |
| Molecular weight | Theoretical: 66.027914 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MSLLKIYWRA MQYLAVERTA TITMCVASVL VALVTLAEPV LFGRVIQSIS DKGDIFSPLL MWAALGGFNI MAAVFVARGA DRLAHRRRL GVMIDSYERL ITMPLAWHQK RGTSNALHTL IRATDSLFTL WLEFMRQHLT TVVALATLIP VAMTMDMRMS L VLIVLGVI ...String: MSLLKIYWRA MQYLAVERTA TITMCVASVL VALVTLAEPV LFGRVIQSIS DKGDIFSPLL MWAALGGFNI MAAVFVARGA DRLAHRRRL GVMIDSYERL ITMPLAWHQK RGTSNALHTL IRATDSLFTL WLEFMRQHLT TVVALATLIP VAMTMDMRMS L VLIVLGVI YVMIGQLVMR KTKDGQAAVE KHHHKLFEHV SDTISNVSVV QSYNRIASET QALRDYAKNL ENAQFPVLNW WA LASGLNR MASTFSMVVV LVLGAYFVTK GQMRVGDVIA FIGFAQLMIG RLDQISAFIN QTVTARAKLE EFFQMEDATA DRQ EPENVA DLNDVKGDIV FDNVTYEFPN SGQGVYDVSF EVKPGQTVAI VGPTGAGKTT LINLLQRVFD PAAGRIMIDG TDTR TVSRR SLRHAIATVF QDAGLFNRSV EDNIRVGRAN ATHEEVHAAA KAAAAHDFIL AKSEGYDTFV GERGSQLSGG ERQRL AIAR AILKDSPILV LDEATSALDV ETEEKVTQAV DELSHNRTTF IIAHRLSTVR SADLVLFMDK GHLVESGSFN ELAERG GRF SDLLRAGGLK LEDKQPKQPV VEGSNVMPFP VKGAVA |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
| Details | abc transporter CBG complex |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 2.5 µm / Calibrated defocus min: 0.8 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.8 µm |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average electron dose: 60.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 180 |
|---|---|
| Output model |  PDB-7zoa: |
 Movie
Movie Controller
Controller







 Z
Z Y
Y X
X