+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
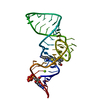
| Title | Ligand bound state of a brocolli-pepper aptamer FRET tile | |||||||||
 Map data Map data | Map sharpened with b-factor of 160 | |||||||||
 Sample Sample |
| |||||||||
| Biological species | synthetic construct (others) | |||||||||
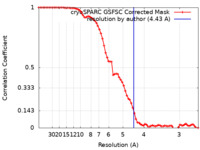
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.43 Å cryo EM / Resolution: 4.43 Å | |||||||||
 Authors Authors | McRae EKS / Vallina NS / Hansen BK / Boussebayle A / Andersen ES | |||||||||
| Funding support |  Denmark, 1 items Denmark, 1 items
| |||||||||
 Citation Citation |  Journal: Acta Crystallogr D Struct Biol / Year: 2018 Journal: Acta Crystallogr D Struct Biol / Year: 2018Title: ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Authors: Tristan Ian Croll /  Abstract: This paper introduces ISOLDE, a new software package designed to provide an intuitive environment for high-fidelity interactive remodelling/refinement of macromolecular models into electron-density ...This paper introduces ISOLDE, a new software package designed to provide an intuitive environment for high-fidelity interactive remodelling/refinement of macromolecular models into electron-density maps. ISOLDE combines interactive molecular-dynamics flexible fitting with modern molecular-graphics visualization and established structural biology libraries to provide an immersive interface wherein the model constantly acts to maintain physically realistic conformations as the user interacts with it by directly tugging atoms with a mouse or haptic interface or applying/removing restraints. In addition, common validation tasks are accelerated and visualized in real time. Using the recently described 3.8 Å resolution cryo-EM structure of the eukaryotic minichromosome maintenance (MCM) helicase complex as a case study, it is demonstrated how ISOLDE can be used alongside other modern refinement tools to avoid common pitfalls of low-resolution modelling and improve the quality of the final model. A detailed analysis of changes between the initial and final model provides a somewhat sobering insight into the dangers of relying on a small number of validation metrics to judge the quality of a low-resolution model. #1:  Journal: Acta Crystallogr D Struct Biol / Year: 2018 Journal: Acta Crystallogr D Struct Biol / Year: 2018Title: Rea-space refinement in PHENIX for cryo-EM and crystallography. Authors: Afonine PV / Poon BK / Read RJ / Sobolev OV / Terwilliger TC / Urzhumtsev A / Adams PD #3:  Journal: Nat Methods / Year: 2017 Journal: Nat Methods / Year: 2017Title: cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Authors: Punjani A / Rubinstein JL / Fleet DJ / Brubaker MA | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14740.map.gz emd_14740.map.gz | 36.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14740-v30.xml emd-14740-v30.xml emd-14740.xml emd-14740.xml | 23.1 KB 23.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14740_fsc.xml emd_14740_fsc.xml | 7.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_14740.png emd_14740.png | 29.6 KB | ||
| Masks |  emd_14740_msk_1.map emd_14740_msk_1.map | 38.4 MB |  Mask map Mask map | |
| Others |  emd_14740_additional_1.map.gz emd_14740_additional_1.map.gz emd_14740_half_map_1.map.gz emd_14740_half_map_1.map.gz emd_14740_half_map_2.map.gz emd_14740_half_map_2.map.gz | 19.2 MB 35.7 MB 35.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14740 http://ftp.pdbj.org/pub/emdb/structures/EMD-14740 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14740 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14740 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14740.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14740.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map sharpened with b-factor of 160 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.29 Å | ||||||||||||||||||||||||||||||||||||
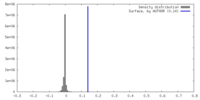
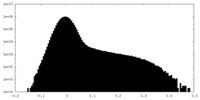
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14740_msk_1.map emd_14740_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
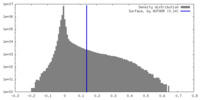
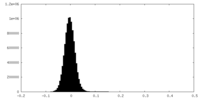
| Density Histograms |
-Additional map: Unsharpened map
| File | emd_14740_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
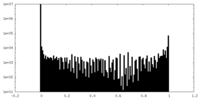
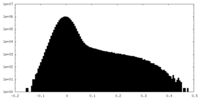
| Density Histograms |
-Half map: Half map B
| File | emd_14740_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
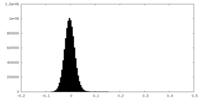
| Density Histograms |
-Half map: Half map A
| File | emd_14740_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ligand bound state of a brocolli-pepper aptamer FRET tile
| Entire | Name: Ligand bound state of a brocolli-pepper aptamer FRET tile |
|---|---|
| Components |
|
-Supramolecule #1: Ligand bound state of a brocolli-pepper aptamer FRET tile
| Supramolecule | Name: Ligand bound state of a brocolli-pepper aptamer FRET tile type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 120 KDa |
-Macromolecule #1: brocolli-pepper aptamer
| Macromolecule | Name: brocolli-pepper aptamer / type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 120.700953 KDa |
| Sequence | String: GGAUACGUCU ACGCUCAGUG ACGGACUCUC UUCGGAGAGU CUGACAUCCG AACCAUACAC GGAUGUGCCU CGCCGAACAG UCUACGGCG AGCUUAAGCG CUGGGGACGC CCAACGCAUC ACAAAGACUG AGUGAUGAAC CAGAAGUAUG GACUGGUUGC G UUGGUGGA ...String: GGAUACGUCU ACGCUCAGUG ACGGACUCUC UUCGGAGAGU CUGACAUCCG AACCAUACAC GGAUGUGCCU CGCCGAACAG UCUACGGCG AGCUUAAGCG CUGGGGACGC CCAACGCAUC ACAAAGACUG AGUGAUGAAC CAGAAGUAUG GACUGGUUGC G UUGGUGGA GACGGUCGGG UCCAGUUCGC UGUCGAGUAG AGUGUGGGCU CCAUCGACGC CGCUUUAAGG UCCCCAAUCG UG GCGUGUC GGCCUGCUUC GGCAGGCACU GGCGCCGGGA CCUUGAAGAG AUGAGAUUUC GAUCUCAUCU UUGGGUGUCU CUG GUGCUU GAGGGCCCUG UGUUCGCACA GGGCCGCUCA CUGGGUGUGG ACGUAUCC |
-Macromolecule #2: 4-[(~{Z})-1-cyano-2-[5-[2-hydroxyethyl(methyl)amino]thieno[3,2-b]...
| Macromolecule | Name: 4-[(~{Z})-1-cyano-2-[5-[2-hydroxyethyl(methyl)amino]thieno[3,2-b]thiophen-2-yl]ethenyl]benzenecarbonitrile type: ligand / ID: 2 / Number of copies: 1 / Formula: J93 |
|---|---|
| Molecular weight | Theoretical: 365.472 Da |
| Chemical component information | 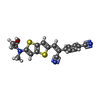 ChemComp-J93: |
-Macromolecule #3: 4-(3,5-difluoro-4-hydroxybenzyl)-1,2-dimethyl-1H-imidazol-5-ol
| Macromolecule | Name: 4-(3,5-difluoro-4-hydroxybenzyl)-1,2-dimethyl-1H-imidazol-5-ol type: ligand / ID: 3 / Number of copies: 1 / Formula: 1TU |
|---|---|
| Molecular weight | Theoretical: 254.233 Da |
| Chemical component information | 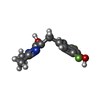 ChemComp-1TU: |
-Macromolecule #4: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 40mM HEPES pH 7.5, 5mM MgCl2, 50mM KCl. Filtered through 0.22 um filter. |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: OTHER / Pretreatment - Pressure: 0.00045000000000000004 kPa / Details: 15mA of current. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 288 K / Instrument: LEICA EM GP Details: 3 uL sample, blotted onto double layer of whatman filter paper for 6 seconds.. |
| Details | Sample was purified by size exclusion chromatography. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.7000000000000001 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.7000000000000001 µm |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 5354 / Average electron dose: 60.0 e/Å2 Details: Collected with a calibrated pixel size of 0.647 Angstrom |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)