[English] 日本語
 Yorodumi
Yorodumi- EMDB-14725: Structure of human OCT3 in complex with inhibitor Corticosterone -
+ Open data
Open data
- Basic information
Basic information
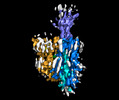
| Entry | 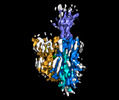 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human OCT3 in complex with inhibitor Corticosterone | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationhistamine transport / epinephrine uptake / histamine metabolic process / norepinephrine uptake / monocarboxylic acid transport / histamine uptake / serotonin transport / spermidine transmembrane transporter activity / quaternary ammonium group transmembrane transporter activity / quaternary ammonium group transport ...histamine transport / epinephrine uptake / histamine metabolic process / norepinephrine uptake / monocarboxylic acid transport / histamine uptake / serotonin transport / spermidine transmembrane transporter activity / quaternary ammonium group transmembrane transporter activity / quaternary ammonium group transport / epinephrine transport / purine-containing compound transmembrane transport / spermidine transport /  organic cation transport / organic cation transport /  Organic cation transport / organic cation transmembrane transporter activity / dopamine uptake / Organic cation transport / organic cation transmembrane transporter activity / dopamine uptake /  regulation of appetite / norepinephrine transport / : / neurotransmitter transmembrane transporter activity / monoamine transmembrane transporter activity / serotonin uptake / toxin transmembrane transporter activity / monoamine transport / Abacavir transmembrane transport / dopamine transport / organic anion transport / organic anion transmembrane transporter activity / regulation of appetite / norepinephrine transport / : / neurotransmitter transmembrane transporter activity / monoamine transmembrane transporter activity / serotonin uptake / toxin transmembrane transporter activity / monoamine transport / Abacavir transmembrane transport / dopamine transport / organic anion transport / organic anion transmembrane transporter activity /  neurotransmitter transport / nuclear outer membrane / xenobiotic transport / transport across blood-brain barrier / neurotransmitter transport / nuclear outer membrane / xenobiotic transport / transport across blood-brain barrier /  endomembrane system / monoatomic ion transport / endomembrane system / monoatomic ion transport /  mitochondrial membrane / presynapse / basolateral plasma membrane / apical plasma membrane / neuronal cell body / mitochondrial membrane / presynapse / basolateral plasma membrane / apical plasma membrane / neuronal cell body /  membrane / membrane /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
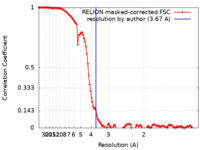
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.67 Å cryo EM / Resolution: 3.67 Å | |||||||||
 Authors Authors | Khanppnavar B / Korkhov V | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural basis of organic cation transporter-3 inhibition. Authors: Basavraj Khanppnavar / Julian Maier / Freja Herborg / Ralph Gradisch / Erika Lazzarin / Dino Luethi / Jae-Won Yang / Chao Qi / Marion Holy / Kathrin Jäntsch / Oliver Kudlacek / Klaus ...Authors: Basavraj Khanppnavar / Julian Maier / Freja Herborg / Ralph Gradisch / Erika Lazzarin / Dino Luethi / Jae-Won Yang / Chao Qi / Marion Holy / Kathrin Jäntsch / Oliver Kudlacek / Klaus Schicker / Thomas Werge / Ulrik Gether / Thomas Stockner / Volodymyr M Korkhov / Harald H Sitte /    Abstract: Organic cation transporters (OCTs) facilitate the translocation of catecholamines, drugs and xenobiotics across the plasma membrane in various tissues throughout the human body. OCT3 plays a key role ...Organic cation transporters (OCTs) facilitate the translocation of catecholamines, drugs and xenobiotics across the plasma membrane in various tissues throughout the human body. OCT3 plays a key role in low-affinity, high-capacity uptake of monoamines in most tissues including heart, brain and liver. Its deregulation plays a role in diseases. Despite its importance, the structural basis of OCT3 function and its inhibition has remained enigmatic. Here we describe the cryo-EM structure of human OCT3 at 3.2 Å resolution. Structures of OCT3 bound to two inhibitors, corticosterone and decynium-22, define the ligand binding pocket and reveal common features of major facilitator transporter inhibitors. In addition, we relate the functional characteristics of an extensive collection of previously uncharacterized human genetic variants to structural features, thereby providing a basis for understanding the impact of OCT3 polymorphisms. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14725.map.gz emd_14725.map.gz | 9.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14725-v30.xml emd-14725-v30.xml emd-14725.xml emd-14725.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14725_fsc.xml emd_14725_fsc.xml | 12.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_14725.png emd_14725.png | 72.5 KB | ||
| Others |  emd_14725_half_map_1.map.gz emd_14725_half_map_1.map.gz emd_14725_half_map_2.map.gz emd_14725_half_map_2.map.gz | 152.3 MB 152.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14725 http://ftp.pdbj.org/pub/emdb/structures/EMD-14725 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14725 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14725 | HTTPS FTP |
-Related structure data
| Related structure data |  7zh6MC  7zh0C  7zhaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14725.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14725.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.66 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half1
| File | emd_14725_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half1 | ||||||||||||
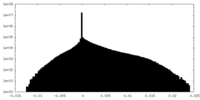
| Projections & Slices |
| ||||||||||||
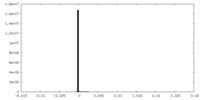
| Density Histograms |
-Half map: half2
| File | emd_14725_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half2 | ||||||||||||
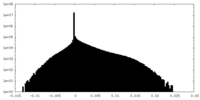
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : human OCT3 in complex with inhibitor Corticosterone
| Entire | Name: human OCT3 in complex with inhibitor Corticosterone |
|---|---|
| Components |
|
-Supramolecule #1: human OCT3 in complex with inhibitor Corticosterone
| Supramolecule | Name: human OCT3 in complex with inhibitor Corticosterone / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier family 22 member 3
| Macromolecule | Name: Solute carrier family 22 member 3 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 61.332836 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPSFDEALQR VGEFGRFQRR VFLLLCLTGV TFAFLFVGVV FLGTQPDHYW CRGPSAAALA ERCGWSPEEE WNRTAPASRG PEPPERRGR CQRYLLEAAN DSASATSALS CADPLAAFPN RSAPLVPCRG GWRYAQAHST IVSEFDLVCV NAWMLDLTQA I LNLGFLTG ...String: MPSFDEALQR VGEFGRFQRR VFLLLCLTGV TFAFLFVGVV FLGTQPDHYW CRGPSAAALA ERCGWSPEEE WNRTAPASRG PEPPERRGR CQRYLLEAAN DSASATSALS CADPLAAFPN RSAPLVPCRG GWRYAQAHST IVSEFDLVCV NAWMLDLTQA I LNLGFLTG AFTLGYAADR YGRIVIYLLS CLGVGVTGVV VAFAPNFPVF VIFRFLQGVF GKGTWMTCYV IVTEIVGSKQ RR IVGIVIQ MFFTLGIIIL PGIAYFIPNW QGIQLAITLP SFLFLLYYWV VPESPRWLIT RKKGDKALQI LRRIAKCNGK YLS SNYSEI TVTDEEVSNP SFLDLVRTPQ MRKCTLILMF AWFTSAVVYQ GLVMRLGIIG GNLYIDFFIS GVVELPGALL ILLT IERLG RRLPFAASNI VAGVACLVTA FLPEGIAWLR TTVATLGRLG ITMAFEIVYL VNSELYPTTL RNFGVSLCSG LCDFG GIIA PFLLFRLAAV WLELPLIIFG ILASICGGLV MLLPETKGIA LPETVDDVEK LGSPHSCKCG RNKKTPVSRS HL |
-Macromolecule #2: CORTICOSTERONE
| Macromolecule | Name: CORTICOSTERONE / type: ligand / ID: 2 / Number of copies: 1 / Formula: C0R |
|---|---|
| Molecular weight | Theoretical: 346.461 Da |
| Chemical component information | 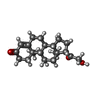 ChemComp-C0R: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm |
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / #0 - Average electron dose: 49.0 e/Å2 / #0 - Details: Dataset 1 / #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / #1 - Average electron dose: 51.0 e/Å2 / #1 - Details: Dataset 2 / #2 - Image recording ID: 3 / #2 - Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / #2 - Average electron dose: 61.0 e/Å2 / #2 - Details: Dataset 3 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



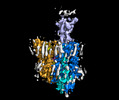



 Z
Z Y
Y X
X