[English] 日本語
 Yorodumi
Yorodumi- EMDB-14355: Structure of the Mimivirus genomic fibre in its relaxed 5-start h... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Mimivirus genomic fibre in its relaxed 5-start helix form | ||||||||||||||||||||||||
 Map data Map data | Masked postprocessed map | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationoxidoreductase activity, acting on CH-OH group of donors /  flavin adenine dinucleotide binding flavin adenine dinucleotide bindingSimilarity search - Function | ||||||||||||||||||||||||
| Biological species |    Acanthamoeba polyphaga mimivirus Acanthamoeba polyphaga mimivirus | ||||||||||||||||||||||||
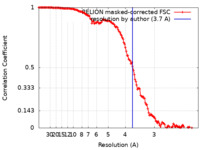
| Method | helical reconstruction /  cryo EM / Resolution: 3.7 Å cryo EM / Resolution: 3.7 Å | ||||||||||||||||||||||||
 Authors Authors | Villalta A / Schmitt A / Estrozi LF / Quemin ERJ / Alempic JM / Lartigue A / Prazak V / Belmudes L / Vasishtan D / Colmant AMG ...Villalta A / Schmitt A / Estrozi LF / Quemin ERJ / Alempic JM / Lartigue A / Prazak V / Belmudes L / Vasishtan D / Colmant AMG / Honore FA / Coute Y / Grunewald K / Abergel C | ||||||||||||||||||||||||
| Funding support |  France, 7 items France, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2022 Journal: Elife / Year: 2022Title: The giant mimivirus 1.2 Mb genome is elegantly organized into a 30-nm diameter helical protein shield. Authors: Alejandro Villalta / Alain Schmitt / Leandro F Estrozi / Emmanuelle R J Quemin / Jean-Marie Alempic / Audrey Lartigue / Vojtěch Pražák / Lucid Belmudes / Daven Vasishtan / Agathe M G ...Authors: Alejandro Villalta / Alain Schmitt / Leandro F Estrozi / Emmanuelle R J Quemin / Jean-Marie Alempic / Audrey Lartigue / Vojtěch Pražák / Lucid Belmudes / Daven Vasishtan / Agathe M G Colmant / Flora A Honoré / Yohann Couté / Kay Grünewald / Chantal Abergel /    Abstract: Mimivirus is the prototype of the family of giant dsDNA viruses. Little is known about the organization of the 1.2 Mb genome inside the membrane-limited nucleoid filling the ~0.5 µm icosahedral ...Mimivirus is the prototype of the family of giant dsDNA viruses. Little is known about the organization of the 1.2 Mb genome inside the membrane-limited nucleoid filling the ~0.5 µm icosahedral capsids. Cryo-electron microscopy, cryo-electron tomography, and proteomics revealed that it is encased into a ~30-nm diameter helical protein shell surprisingly composed of two GMC-type oxidoreductases, which also form the glycosylated fibrils decorating the capsid. The genome is arranged in 5- or 6-start left-handed super-helices, with each DNA-strand lining the central channel. This luminal channel of the nucleoprotein fiber is wide enough to accommodate oxidative stress proteins and RNA polymerase subunits identified by proteomics. Such elegant supramolecular organization would represent a remarkable evolutionary strategy for packaging and protecting the genome, in a state ready for immediate transcription upon unwinding in the host cytoplasm. The parsimonious use of the same protein in two unrelated substructures of the virion is unexpected for a giant virus with thousand genes at its disposal. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14355.map.gz emd_14355.map.gz | 34.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14355-v30.xml emd-14355-v30.xml emd-14355.xml emd-14355.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14355_fsc.xml emd_14355_fsc.xml | 14.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_14355.png emd_14355.png | 84.4 KB | ||
| Others |  emd_14355_additional_1.map.gz emd_14355_additional_1.map.gz emd_14355_half_map_1.map.gz emd_14355_half_map_1.map.gz emd_14355_half_map_2.map.gz emd_14355_half_map_2.map.gz | 193.1 MB 193.6 MB 194 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14355 http://ftp.pdbj.org/pub/emdb/structures/EMD-14355 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14355 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14355 | HTTPS FTP |
-Related structure data
| Related structure data |  7yx5MC  7ptvC  7yx3C  7yx4C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14355.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14355.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Masked postprocessed map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||
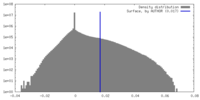
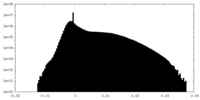
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unmasked map before postprocessing
| File | emd_14355_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unmasked map before postprocessing | ||||||||||||
| Projections & Slices |
| ||||||||||||
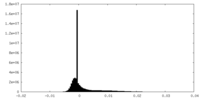
| Density Histograms |
-Half map: Unmasked half map
| File | emd_14355_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unmasked half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
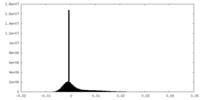
| Density Histograms |
-Half map: Unmasked half map
| File | emd_14355_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unmasked half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Mimivirus genomic fibre in its relaxed 5-start helix form
| Entire | Name: Mimivirus genomic fibre in its relaxed 5-start helix form |
|---|---|
| Components |
|
-Supramolecule #1: Mimivirus genomic fibre in its relaxed 5-start helix form
| Supramolecule | Name: Mimivirus genomic fibre in its relaxed 5-start helix form type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:    Acanthamoeba polyphaga mimivirus / Strain: Reunion Acanthamoeba polyphaga mimivirus / Strain: Reunion |
-Macromolecule #1: Putative glucose-methanol-choline oxidoreductase protein
| Macromolecule | Name: Putative glucose-methanol-choline oxidoreductase protein type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Acanthamoeba polyphaga mimivirus Acanthamoeba polyphaga mimivirus |
| Molecular weight | Theoretical: 76.431055 KDa |
| Sequence | String: MAHRSRCNCN DTSNSNGSQH GINLPLRKID TYDPCVNCRV KPHLCPKPHP CPKPENLEAD IVIIGAGAAG CVLAYYLTKF SDLKIILLE AGHTHFNDPV VTDPMGFFGK YNPPNENIRM SQNPSYAWQP ALEPDTGAYS MRNVVAHGLA VGGSTAINQL N YIVGGRTV ...String: MAHRSRCNCN DTSNSNGSQH GINLPLRKID TYDPCVNCRV KPHLCPKPHP CPKPENLEAD IVIIGAGAAG CVLAYYLTKF SDLKIILLE AGHTHFNDPV VTDPMGFFGK YNPPNENIRM SQNPSYAWQP ALEPDTGAYS MRNVVAHGLA VGGSTAINQL N YIVGGRTV FDNDWPTGWK YDDIKKYFRR VLADISPIRD GTKVNLTNTI LESMRVLADQ QVSSGVPVDF LINKATGGLP NI EQTYQGA PIVNLNDYEG INSVCGFKSY YVGVNQLSDG SYIRKYAGNT YLNSYYVDSN GFGIGKFSNL RVISDAVVDR IHF EGQRAV SVTYIDKKGN LHSVKVHKEV EICSGSFFTP TILQRSGIGD FSYLSSIGVP DLVYNNPLVG QGLRNHYSPI TQVS VTGPD AAAFLSNTAA GPTNMSFRGA GMLGYHKLEP NKPSNAGSVT YRKYELLVTG GVAISADQQY LSGISSSTGN YFALI ADDI RFAPVGYIKI GTPNFPRDTP KIFFNTFVNY TPTTDPADQQ WPVAQKTLAP LISALLGYDA IYQIVQQMKV VAVNAG FNV TLQMAYPPND LLVELHNGLN TYGINWWHYF VPSLVNDDTP AGKLFASTLS KLSYYPRSGA HLDSHQSCSC SIGGTVD TE LKVIGVENVR VTDLSAAPHP PGGNTWCTAA MIGARATDLI LGKPLVANLP PEDVPVFTTS |
-Macromolecule #2: FLAVIN-ADENINE DINUCLEOTIDE
| Macromolecule | Name: FLAVIN-ADENINE DINUCLEOTIDE / type: ligand / ID: 2 / Number of copies: 1 / Formula: FAD |
|---|---|
| Molecular weight | Theoretical: 785.55 Da |
| Chemical component information |  ChemComp-FAD: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Component - Concentration: 40.0 mM / Component - Formula: (HOCH2)3CNH2 / Component - Name: Tris buffer |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 50.6 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)