[English] 日本語
 Yorodumi
Yorodumi- EMDB-14332: 1.58 A STRUCTURE OF HUMAN APOFERRITIN OBTAINED FROM TITAN KRIOS 2... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 1.58 A STRUCTURE OF HUMAN APOFERRITIN OBTAINED FROM TITAN KRIOS 2 AT eBIC, DLS UNDER COMMISSIONING SESSION CM26464-2 | |||||||||
 Map data Map data | ApoF structure post processed in Relion | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Protein standard / METAL BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationiron ion sequestering activity / Scavenging by Class A Receptors / intracellular ferritin complex /  autolysosome / Golgi Associated Vesicle Biogenesis / autolysosome / Golgi Associated Vesicle Biogenesis /  ferroxidase / intracellular sequestering of iron ion / ferroxidase / intracellular sequestering of iron ion /  ferroxidase activity / negative regulation of fibroblast proliferation / ferroxidase activity / negative regulation of fibroblast proliferation /  ferric iron binding ...iron ion sequestering activity / Scavenging by Class A Receptors / intracellular ferritin complex / ferric iron binding ...iron ion sequestering activity / Scavenging by Class A Receptors / intracellular ferritin complex /  autolysosome / Golgi Associated Vesicle Biogenesis / autolysosome / Golgi Associated Vesicle Biogenesis /  ferroxidase / intracellular sequestering of iron ion / ferroxidase / intracellular sequestering of iron ion /  ferroxidase activity / negative regulation of fibroblast proliferation / ferroxidase activity / negative regulation of fibroblast proliferation /  ferric iron binding / ferric iron binding /  ferrous iron binding / Iron uptake and transport / tertiary granule lumen / iron ion transport / intracellular iron ion homeostasis / ficolin-1-rich granule lumen / ferrous iron binding / Iron uptake and transport / tertiary granule lumen / iron ion transport / intracellular iron ion homeostasis / ficolin-1-rich granule lumen /  immune response / iron ion binding / negative regulation of cell population proliferation / Neutrophil degranulation / extracellular exosome / extracellular region / identical protein binding / immune response / iron ion binding / negative regulation of cell population proliferation / Neutrophil degranulation / extracellular exosome / extracellular region / identical protein binding /  nucleus / nucleus /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 1.58 Å cryo EM / Resolution: 1.58 Å | |||||||||
 Authors Authors | Clare DK | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Faraday Discuss / Year: 2022 Journal: Faraday Discuss / Year: 2022Title: Application of super-resolution and correlative double sampling in cryo-electron microscopy. Authors: Yuewen Sheng / Peter J Harrison / Vinod Vogirala / Zhengyi Yang / Claire Strain-Damerell / Thomas Frosio / Benjamin A Himes / C Alistair Siebert / Peijun Zhang / Daniel K Clare /   Abstract: Developments in cryo-EM have allowed atomic or near-atomic resolution structure determination to become routine in single particle analysis (SPA). However, near-atomic resolution structures ...Developments in cryo-EM have allowed atomic or near-atomic resolution structure determination to become routine in single particle analysis (SPA). However, near-atomic resolution structures determined using cryo-electron tomography and sub-tomogram averaging (cryo-ET STA) are much less routine. In this paper, we show that collecting cryo-ET STA data using the same conditions as SPA, with both correlated double sampling (CDS) and the super-resolution mode, allowed apoferritin to be reconstructed out to the physical Nyquist frequency of the images. Even with just two tilt series, STA yields an apoferritin map at 2.9 Å resolution. These results highlight the exciting potential of cryo-ET STA in the future of protein structure determination. While processing SPA data recorded in super-resolution mode may yield structures surpassing the physical Nyquist limit, processing cryo-ET STA data in the super-resolution mode gave no additional resolution benefit. We further show that collecting SPA data in the super-resolution mode, with CDS activated, reduces the estimated -factor, leading to a reduction in the number of particles required to reach a target resolution without compromising the data size on disk and the area imaged in SerialEM. However, collecting SPA data in CDS does reduce throughput, given that a similar resolution structure, with a slightly larger -factor, is achievable with optimised parameters for speed in EPU (without CDS). | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14332.map.gz emd_14332.map.gz | 58 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14332-v30.xml emd-14332-v30.xml emd-14332.xml emd-14332.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14332_fsc.xml emd_14332_fsc.xml | 21 KB | Display |  FSC data file FSC data file |
| Images |  emd_14332.png emd_14332.png | 321 KB | ||
| Others |  emd_14332_half_map_1.map.gz emd_14332_half_map_1.map.gz emd_14332_half_map_2.map.gz emd_14332_half_map_2.map.gz | 660.5 MB 660.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14332 http://ftp.pdbj.org/pub/emdb/structures/EMD-14332 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14332 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14332 | HTTPS FTP |
-Related structure data
| Related structure data |  7r5oMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14332.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14332.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ApoF structure post processed in Relion | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.656 Å | ||||||||||||||||||||||||||||||||||||
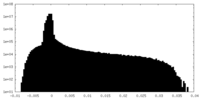
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map
| File | emd_14332_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
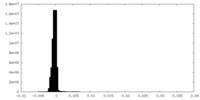
| Density Histograms |
-Half map: Half map
| File | emd_14332_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : human Apoferritin
| Entire | Name: human Apoferritin Ferritin Ferritin |
|---|---|
| Components |
|
-Supramolecule #1: human Apoferritin
| Supramolecule | Name: human Apoferritin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: Heavy chain |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 500 KDa |
-Macromolecule #1: Ferritin heavy chain, N-terminally processed
| Macromolecule | Name: Ferritin heavy chain, N-terminally processed / type: protein_or_peptide / ID: 1 / Number of copies: 24 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 20.21857 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: STSQVRQNYH QDSEAAINRQ INLELYASYV YLSMSYYFDR DDVALKNFAK YFLHQSHEER EHAEKLMKLQ NQRGGRIFLQ DIQKPD(CSX)DD WESGLNAMEC ALHLEKNVNQ SLLELHKLAT DKNDPHLCDF IETHYLNEQV KAIKELGDHV TNLRKMG AP ESGLAEYLFD KHTLG |
-Macromolecule #2: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 2 / Number of copies: 24 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 3384 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM TCEP |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: blot time of 2.5 seconds. |
| Details | The apoferritin sample was applied to in-house graphene-coated Quantifoil r2/2 EM grid |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 130000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 130000 |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 2295 / Average exposure time: 2.5 sec. / Average electron dose: 34.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)