[English] 日本語
 Yorodumi
Yorodumi- EMDB-14178: Fiber-forming RubisCO derived from ancestral sequence reconstruct... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Fiber-forming RubisCO derived from ancestral sequence reconstruction and rational engineering | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species | synthetic construct (others) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.0 Å cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Schulz L / Zarzycki J / Prinz S / Schuller JM / Erb TJ / Hochberg GKA | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Evolution of increased complexity and specificity at the dawn of form I Rubiscos. Authors: Luca Schulz / Zhijun Guo / Jan Zarzycki / Wieland Steinchen / Jan M Schuller / Thomas Heimerl / Simone Prinz / Oliver Mueller-Cajar / Tobias J Erb / Georg K A Hochberg /   Abstract: The evolution of ribulose-1,5-bisphosphate carboxylase/oxygenases (Rubiscos) that discriminate strongly between their substrate carbon dioxide and the undesired side substrate dioxygen was an ...The evolution of ribulose-1,5-bisphosphate carboxylase/oxygenases (Rubiscos) that discriminate strongly between their substrate carbon dioxide and the undesired side substrate dioxygen was an important event for photosynthetic organisms adapting to an oxygenated environment. We use ancestral sequence reconstruction to recapitulate this event. We show that Rubisco increased its specificity and carboxylation efficiency through the gain of an accessory subunit before atmospheric oxygen was present. Using structural and biochemical approaches, we retrace how this subunit was gained and became essential. Our work illuminates the emergence of an adaptation to rising ambient oxygen levels, provides a template for investigating the function of interactions that have remained elusive because of their essentiality, and sheds light on the determinants of specificity in Rubisco. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14178.map.gz emd_14178.map.gz | 230.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14178-v30.xml emd-14178-v30.xml emd-14178.xml emd-14178.xml | 13.5 KB 13.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_14178.png emd_14178.png | 53.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14178 http://ftp.pdbj.org/pub/emdb/structures/EMD-14178 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14178 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14178 | HTTPS FTP |
-Related structure data
| Related structure data |  7qviMC  7qsvC  7qswC  7qsxC  7qsyC  7qszC  7qt1C M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14178.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14178.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.837 Å | ||||||||||||||||||||||||||||||||||||
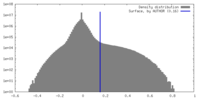
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : ancestral sequence reconstruction of RubisCO large subunit in com...
| Entire | Name: ancestral sequence reconstruction of RubisCO large subunit in complex with CABP and Magnesium |
|---|---|
| Components |
|
-Supramolecule #1: ancestral sequence reconstruction of RubisCO large subunit in com...
| Supramolecule | Name: ancestral sequence reconstruction of RubisCO large subunit in complex with CABP and Magnesium type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: complex of two octamers (2x L8) |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Recombinant expression | Organism:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria)Recombinant strain: ArtricExpress (DE3) |
| Molecular weight | Experimental: 873 KDa |
-Macromolecule #1: RubisCO large subunit
| Macromolecule | Name: RubisCO large subunit / type: protein_or_peptide / ID: 1 / Number of copies: 16 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 51.19693 KDa |
| Recombinant expression | Organism:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) |
| Sequence | String: MARAQYEAGV RPYRETYYDP DYEPKDTDLL CAFRITPKPG VPMEEAAAAV AAESSTGTWT EVWSNLLTDL ERYKARCYRI EGDVAYIAY PLDLFEEGSI VNIMSSIVGN VFGFKAVQAL RLEDMRIPVA YLKTFPGPPT GIQVERDRLN KYGRPLLGGT I KPKLGLSA ...String: MARAQYEAGV RPYRETYYDP DYEPKDTDLL CAFRITPKPG VPMEEAAAAV AAESSTGTWT EVWSNLLTDL ERYKARCYRI EGDVAYIAY PLDLFEEGSI VNIMSSIVGN VFGFKAVQAL RLEDMRIPVA YLKTFPGPPT GIQVERDRLN KYGRPLLGGT I KPKLGLSA KEYARVVYEC LRGGLDTT(KCX)D DENLNSQPFN RWRDRFLYVM EAVRKAEAET GERKGHWLNV TAGSTEEM L KRAEFAAELG SRYIMVDFLT AGFAAFASVR RRAEELGLML HCHRAMHAVF DRQPNHGIHF RVLAKWLRMV GGDHVHTGT VVGKLEGDRA ETLGIADLLR EDYVPADPGR GLFFDQDWAG LKPVFPVASG GIHVWHVPDL VSIFGDDAFF LFGGGTHGHP RGSRAGATA NRVAVEAVVQ ARNEGRDILA EGREILEEAA RWCPELREAM ELWGDVKFEV EA |
-Macromolecule #2: 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE
| Macromolecule | Name: 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 16 / Formula: CAP |
|---|---|
| Molecular weight | Theoretical: 356.115 Da |
| Chemical component information |  ChemComp-CAP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 16 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | 2D array |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R2/1 / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 171572 |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)