+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 S protein S:A222V + S:D614G mutant 2-up | |||||||||||||||||||||||||||
 Map data Map data | SARS-CoV-2 S protein S:A222V S:D614G mutant 2-up | |||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||
 Keywords Keywords |  SARS-CoV-2 / SARS-CoV-2 /  S protein / S:A222V + S:D614G mutant / S protein / S:A222V + S:D614G mutant /  VIRAL PROTEIN VIRAL PROTEIN | |||||||||||||||||||||||||||
| Biological species |   Severe acute respiratory syndrome-related coronavirus Severe acute respiratory syndrome-related coronavirus | |||||||||||||||||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 6.8 Å cryo EM / Resolution: 6.8 Å | |||||||||||||||||||||||||||
 Authors Authors | Ginex T / Marco-Marin C / Wieczor M / Mata CP / Krieger J / Lopez-Redondo ML / Ruiz-Rodriguez P / Melero R / Sanchez-Sorzano CO / Martinez M ...Ginex T / Marco-Marin C / Wieczor M / Mata CP / Krieger J / Lopez-Redondo ML / Ruiz-Rodriguez P / Melero R / Sanchez-Sorzano CO / Martinez M / Gougeard N / Forcada-Nadal A / Zamora-Caballero S / Gozalbo-Rovira R / Sanz-Frasquet C / Bravo J / Rubio V / Marina A / Geller R / Comas I / Gil C / Coscolla M / Orozco M / Llacer JL / Carazo JM | |||||||||||||||||||||||||||
| Funding support |  Spain, 8 items Spain, 8 items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2022 Journal: PLoS Pathog / Year: 2022Title: The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation. Authors: Tiziana Ginex / Clara Marco-Marín / Miłosz Wieczór / Carlos P Mata / James Krieger / Paula Ruiz-Rodriguez / Maria Luisa López-Redondo / Clara Francés-Gómez / Roberto Melero / Carlos ...Authors: Tiziana Ginex / Clara Marco-Marín / Miłosz Wieczór / Carlos P Mata / James Krieger / Paula Ruiz-Rodriguez / Maria Luisa López-Redondo / Clara Francés-Gómez / Roberto Melero / Carlos Óscar Sánchez-Sorzano / Marta Martínez / Nadine Gougeard / Alicia Forcada-Nadal / Sara Zamora-Caballero / Roberto Gozalbo-Rovira / Carla Sanz-Frasquet / Rocío Arranz / Jeronimo Bravo / Vicente Rubio / Alberto Marina / / Ron Geller / Iñaki Comas / Carmen Gil / Mireia Coscolla / Modesto Orozco / José Luis Llácer / Jose-Maria Carazo /  Abstract: The S:A222V point mutation, within the G clade, was characteristic of the 20E (EU1) SARS-CoV-2 variant identified in Spain in early summer 2020. This mutation has since reappeared in the Delta ...The S:A222V point mutation, within the G clade, was characteristic of the 20E (EU1) SARS-CoV-2 variant identified in Spain in early summer 2020. This mutation has since reappeared in the Delta subvariant AY.4.2, raising questions about its specific effect on viral infection. We report combined serological, functional, structural and computational studies characterizing the impact of this mutation. Our results reveal that S:A222V promotes an increased RBD opening and slightly increases ACE2 binding as compared to the parent S:D614G clade. Finally, S:A222V does not reduce sera neutralization capacity, suggesting it does not affect vaccine effectiveness. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13917.map.gz emd_13917.map.gz | 58.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13917-v30.xml emd-13917-v30.xml emd-13917.xml emd-13917.xml | 23.9 KB 23.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13917.png emd_13917.png | 93.8 KB | ||
| Filedesc metadata |  emd-13917.cif.gz emd-13917.cif.gz | 6.8 KB | ||
| Others |  emd_13917_additional_1.map.gz emd_13917_additional_1.map.gz emd_13917_half_map_1.map.gz emd_13917_half_map_1.map.gz emd_13917_half_map_2.map.gz emd_13917_half_map_2.map.gz | 50.1 MB 78.1 MB 78.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13917 http://ftp.pdbj.org/pub/emdb/structures/EMD-13917 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13917 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13917 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13917.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13917.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
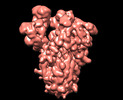
| Annotation | SARS-CoV-2 S protein S:A222V S:D614G mutant 2-up | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.4 Å | ||||||||||||||||||||||||||||||||||||
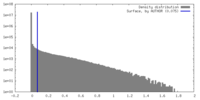
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: SARS-CoV-2 S protein S:A222V S:D614G mutant 2-up unsharpened map
| File | emd_13917_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SARS-CoV-2 S protein S:A222V S:D614G mutant 2-up unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
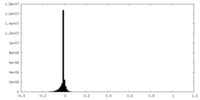
| Density Histograms |
-Half map: SARS-CoV-2 S protein S:A222V S:D614G mutant 2-up half map 1
| File | emd_13917_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SARS-CoV-2 S protein S:A222V S:D614G mutant 2-up half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
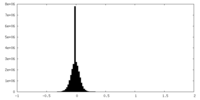
| Density Histograms |
-Half map: SARS-CoV-2 S protein S:A222V S:D614G mutant 2-up half map 2
| File | emd_13917_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SARS-CoV-2 S protein S:A222V S:D614G mutant 2-up half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
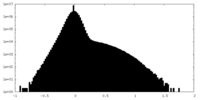
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 S protein S:A222V + S:D614G mutant
| Entire | Name: SARS-CoV-2 S protein S:A222V + S:D614G mutant |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 S protein S:A222V + S:D614G mutant
| Supramolecule | Name: SARS-CoV-2 S protein S:A222V + S:D614G mutant / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Severe acute respiratory syndrome-related coronavirus Severe acute respiratory syndrome-related coronavirus |
| Molecular weight | Theoretical: 340 KDa |
-Macromolecule #1: SARS-CoV-2 S protein S:A222V + S:D614G mutant
| Macromolecule | Name: SARS-CoV-2 S protein S:A222V + S:D614G mutant / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Severe acute respiratory syndrome-related coronavirus Severe acute respiratory syndrome-related coronavirus |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: CVNLTTRTQL PPAYTNSFTR GVYYPDKVFR SSVLHSTQDL FLPFFSNVTW FHAIHVSGTN GTKRFDNPVL PFNDGVYFAS TEKSNIIRGW IFGTTLDSKT QSLLIVNNAT NVVIKVCEFQ FCNDPFLGVY YHKNNKSWME SEFRVYSSAN NCTFEYVSQP FLMDLEGKQG ...String: CVNLTTRTQL PPAYTNSFTR GVYYPDKVFR SSVLHSTQDL FLPFFSNVTW FHAIHVSGTN GTKRFDNPVL PFNDGVYFAS TEKSNIIRGW IFGTTLDSKT QSLLIVNNAT NVVIKVCEFQ FCNDPFLGVY YHKNNKSWME SEFRVYSSAN NCTFEYVSQP FLMDLEGKQG NFKNLREFVF KNIDGYFKIY SKHTPINLVR DLPQGFSVLE PLVDLPIGIN ITRFQTLLAL HRSYLTPGDS SSGWTAGAAA YYVGYLQPRT FLLKYNENGT ITDAVDCALD PLSETKCTLK SFTVEKGIYQ TSNFRVQPTE SIVRFPNITN LCPFGEVFNA TRFASVYAWN RKRISNCVAD YSVLYNSASF STFKCYGVSP TKLNDLCFTN VYADSFVIRG DEVRQIAPGQ TGKIADYNYK LPDDFTGCVI AWNSNNLDSK VGGNYNYLYR LFRKSNLKPF ERDISTEIYQ AGSTPCNGVE GFNCYFPLQS YGFQPTNGVG YQPYRVVVLS FELLHAPATV CGPKKSTNLV KNKCVNFNFN GLTGTGVLTE SNKKFLPFQQ FGRDIADTTD AVRDPQTLEI LDITPCSFGG VSVITPGTNT SNQVAVLYQG VNCTEVPVAI HADQLTPTWR VYSTGSNVFQ TRAGCLIGAE HVNNSYECDI PIGAGICASY QTQTNSPASV ASQSIIAYTM SLGAENSVAY SNNSIAIPTN FTISVTTEIL PVSMTKTSVD CTMYICGDST ECSNLLLQYG SFCTQLNRAL TGIAVEQDKN TQEVFAQVKQ IYKTPPIKDF GGFNFSQILP DPSKPSKRSF IEDLLFNKVT LADAGFIKQY GDCLGDIAAR DLICAQKFNG LTVLPPLLTD EMIAQYTSAL LAGTITSGWT FGAGAALQIP FAMQMAYRFN GIGVTQNVLY ENQKLIANQF NSAIGKIQDS LSSTASALGK LQDVVNQNAQ ALNTLVKQLS SNFGAISSVL NDILSRLDPP EAEVQIDRLI TGRLQSLQTY VTQQLIRAAE IRASANLAAT KMSECVLGQS KRVDFCGKGY HLMSFPQSAP HGVVFLHVTY VPAQEKNFTT APAICHDGKA HFPREGVFVS NGTHWFVTQR NFYEPQIITT DNTFVSGNCD VVIGIVNNTV YDPLQPELDS FKEELDKYFK NHTSPDVDLG DISGINASVV NIQKEIDRLN EVAKNLNESL IDLQELGKYE QYIKWPLVPR GSGYIPEAPR DGQAYVRKDG EWVFLSTFLS PHHHHHHHHH EQKLISEEDL |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.2 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 283 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: OTHER / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.3 µm / Nominal magnification: 120000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 4841 / Average exposure time: 20.0 sec. / Average electron dose: 32.4 e/Å2 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 509466 |
|---|---|
| Startup model | Type of model: OTHER / Details: Initial model generation |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC |
| Final 3D classification | Software: (Name: RELION, Scipion) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: FOURIER SPACE / Resolution.type: BY AUTHOR / Resolution: 6.8 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 4808 |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)