[English] 日本語
 Yorodumi
Yorodumi- EMDB-13752: Human GYS1-GYG1 complex activated state bound to glucose-6-phosphate -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human GYS1-GYG1 complex activated state bound to glucose-6-phosphate | |||||||||
 Map data Map data | Locally filtered map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationglycogen synthase activity, transferring glucose-1-phosphate / Glycogen storage disease type XV (GYG1) / Glycogen storage disease type 0 (muscle GYS1) /  glycogen(starch) synthase / Glycogen storage disease type II (GAA) / glycogen(starch) synthase / Glycogen storage disease type II (GAA) /  glycogenin glucosyltransferase / glycogenin glucosyltransferase /  glycogenin glucosyltransferase activity / UDP-alpha-D-glucose:glucosyl-glycogenin alpha-D-glucosyltransferase activity / glycogen (starch) synthase activity / glycogenin glucosyltransferase activity / UDP-alpha-D-glucose:glucosyl-glycogenin alpha-D-glucosyltransferase activity / glycogen (starch) synthase activity /  glucose binding ...glycogen synthase activity, transferring glucose-1-phosphate / Glycogen storage disease type XV (GYG1) / Glycogen storage disease type 0 (muscle GYS1) / glucose binding ...glycogen synthase activity, transferring glucose-1-phosphate / Glycogen storage disease type XV (GYG1) / Glycogen storage disease type 0 (muscle GYS1) /  glycogen(starch) synthase / Glycogen storage disease type II (GAA) / glycogen(starch) synthase / Glycogen storage disease type II (GAA) /  glycogenin glucosyltransferase / glycogenin glucosyltransferase /  glycogenin glucosyltransferase activity / UDP-alpha-D-glucose:glucosyl-glycogenin alpha-D-glucosyltransferase activity / glycogen (starch) synthase activity / glycogenin glucosyltransferase activity / UDP-alpha-D-glucose:glucosyl-glycogenin alpha-D-glucosyltransferase activity / glycogen (starch) synthase activity /  glucose binding / glycogen biosynthetic process / Glycogen breakdown (glycogenolysis) / glucose binding / glycogen biosynthetic process / Glycogen breakdown (glycogenolysis) /  glycosyltransferase activity / glycosyltransferase activity /  inclusion body / Myoclonic epilepsy of Lafora / inclusion body / Myoclonic epilepsy of Lafora /  Glycogen synthesis / lysosomal lumen / manganese ion binding / Glycogen synthesis / lysosomal lumen / manganese ion binding /  heart development / secretory granule lumen / ficolin-1-rich granule lumen / Neutrophil degranulation / protein homodimerization activity / extracellular region / heart development / secretory granule lumen / ficolin-1-rich granule lumen / Neutrophil degranulation / protein homodimerization activity / extracellular region /  membrane / membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.7 Å cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | McCorvie TJ / Shrestha L / Froese DS / Ferreira IM / Yue WW | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Molecular basis for the regulation of human glycogen synthase by phosphorylation and glucose-6-phosphate. Authors: Thomas J McCorvie / Paula M Loria / Meihua Tu / Seungil Han / Leela Shrestha / D Sean Froese / Igor M Ferreira / Allison P Berg / Wyatt W Yue /    Abstract: Glycogen synthase (GYS1) is the central enzyme in muscle glycogen biosynthesis. GYS1 activity is inhibited by phosphorylation of its amino (N) and carboxyl (C) termini, which is relieved by ...Glycogen synthase (GYS1) is the central enzyme in muscle glycogen biosynthesis. GYS1 activity is inhibited by phosphorylation of its amino (N) and carboxyl (C) termini, which is relieved by allosteric activation of glucose-6-phosphate (Glc6P). We present cryo-EM structures at 3.0-4.0 Å resolution of phosphorylated human GYS1, in complex with a minimal interacting region of glycogenin, in the inhibited, activated and catalytically competent states. Phosphorylations of specific terminal residues are sensed by different arginine clusters, locking the GYS1 tetramer in an inhibited state via intersubunit interactions. The Glc6P activator promotes conformational change by disrupting these interactions and increases the flexibility of GYS1, such that it is poised to adopt a catalytically competent state when the sugar donor UDP-glucose (UDP-glc) binds. We also identify an inhibited-like conformation that has not transitioned into the activated state, in which the locking interaction of phosphorylation with the arginine cluster impedes subsequent conformational changes due to Glc6P binding. Our results address longstanding questions regarding the mechanism of human GYS1 regulation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13752.map.gz emd_13752.map.gz | 62.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13752-v30.xml emd-13752-v30.xml emd-13752.xml emd-13752.xml | 26.6 KB 26.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13752_fsc.xml emd_13752_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_13752.png emd_13752.png | 166.1 KB | ||
| Others |  emd_13752_additional_1.map.gz emd_13752_additional_1.map.gz emd_13752_additional_2.map.gz emd_13752_additional_2.map.gz emd_13752_half_map_1.map.gz emd_13752_half_map_1.map.gz emd_13752_half_map_2.map.gz emd_13752_half_map_2.map.gz | 8.3 MB 79.1 MB 79.6 MB 79.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13752 http://ftp.pdbj.org/pub/emdb/structures/EMD-13752 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13752 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13752 | HTTPS FTP |
-Related structure data
| Related structure data |  7q12MC  7q0bC  7q0sC  7q13C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13752.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13752.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Locally filtered map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.086 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Sharpened map
| File | emd_13752_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Non-sharpened map
| File | emd_13752_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Non-sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 1
| File | emd_13752_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 2
| File | emd_13752_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of phosphorylated full-length human glycogen synt...
| Entire | Name: Ternary complex of phosphorylated full-length human glycogen synthase 1 in complex with minimum region of human glycogenin-1 |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of phosphorylated full-length human glycogen synt...
| Supramolecule | Name: Ternary complex of phosphorylated full-length human glycogen synthase 1 in complex with minimum region of human glycogenin-1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / Tissue: muscle Homo sapiens (human) / Tissue: muscle |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Molecular weight | Theoretical: 350 KDa |
-Macromolecule #1: Glycogen [starch] synthase, muscle
| Macromolecule | Name: Glycogen [starch] synthase, muscle / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number:  glycogen(starch) synthase glycogen(starch) synthase |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / Tissue: Muscle Homo sapiens (human) / Tissue: Muscle |
| Molecular weight | Theoretical: 83.88543 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MPLNRTLSMS SLPGLEDWED EFDLENAVLF EVAWEVANKV GGIYTVLQTK AKVTGDEWGD NYFLVGPYTE QGVRTQVELL EAPTPALKR TLDSMNSKGC KVYFGRWLIE GGPLVVLLDV GASAWALERW KGELWDTCNI GVPWYDREAN DAVLFGFLTT W FLGEFLAQ ...String: MPLNRTLSMS SLPGLEDWED EFDLENAVLF EVAWEVANKV GGIYTVLQTK AKVTGDEWGD NYFLVGPYTE QGVRTQVELL EAPTPALKR TLDSMNSKGC KVYFGRWLIE GGPLVVLLDV GASAWALERW KGELWDTCNI GVPWYDREAN DAVLFGFLTT W FLGEFLAQ SEEKPHVVAH FHEWLAGVGL CLCRARRLPV ATIFTTHATL LGRYLCAGAV DFYNNLENFN VDKEAGERQI YH RYCMERA AAHCAHVFTT VSQITAIEAQ HLLKRKPDIV TPNGLNVKKF SAMHEFQNLH AQSKARIQEF VRGHFYGHLD FNL DKTLYF FIAGRYEFSN KGADVFLEAL ARLNYLLRVN GSEQTVVAFF IMPARTNNFN VETLKGQAVR KQLWDTANTV KEKF GRKLY ESLLVGSLPD MNKMLDKEDF TMMKRAIFAT QRQSFPPVCT HNMLDDSSDP ILTTIRRIGL FNSSADRVKV IFHPE FLSS TSPLLPVDYE EFVRGCHLGV FPSYYEPWGY TPAECTVMGI PSISTNLSGF GCFMEEHIAD PSAYGIYILD RRFRSL DDS CSQLTSFLYS FCQQSRRQRI IQRNRTERLS DLLDWKYLGR YYMSARHMAL SKAFPEHFTY EPNEADAAQG YRYPRPA SV PPSPSLSRHS SPHQSEDEED PRNGPLEEDG ERYDEDEEAA KDRRNIRAPE WPRRASCTSS TSGSKRNSVD TATSSSLS T PSEPLSPTSS LGEERN |
-Macromolecule #2: Glycogenin-1
| Macromolecule | Name: Glycogenin-1 / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO / EC number:  glycogenin glucosyltransferase glycogenin glucosyltransferase |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.422621 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MTDQAFVTLT TNDAYAKGAL VLGSSLKQHR TTRRLVVLAT PQVSDSMRKV LETVFDEVIM VDVLDSGDSA HLTLMKRPEL GVTLTKLHC WSLTQYSKCV FMDADTLVLA NIDDLFDREE LSAAPDPGWP DCFNSGVFVY QPSVETYNQL LHLASEQGSF D GGDQGILN ...String: MTDQAFVTLT TNDAYAKGAL VLGSSLKQHR TTRRLVVLAT PQVSDSMRKV LETVFDEVIM VDVLDSGDSA HLTLMKRPEL GVTLTKLHC WSLTQYSKCV FMDADTLVLA NIDDLFDREE LSAAPDPGWP DCFNSGVFVY QPSVETYNQL LHLASEQGSF D GGDQGILN TFFSSWATTD IRKHLPFIYN LSSISIYSYL PAFKVFGASA KVVHFLGRVK PWNYTYDPKT KSVKSEAHDP NM THPEFLI LWWNIFTTNV LPLLQQFGLV KDTCSYVNVL SDLVYTLAFS CGFCRKEDVS GAISHLSLGE IPAMAQPFVS SEE RKERWE QGQADYMGAD SFDNIKRKLD TYLQ |
-Macromolecule #3: 6-O-phosphono-alpha-D-glucopyranose
| Macromolecule | Name: 6-O-phosphono-alpha-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 4 / Formula: G6P |
|---|---|
| Molecular weight | Theoretical: 260.136 Da |
| Chemical component information | 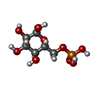 ChemComp-G6P: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.75 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: 25 mM HEPES, pH 7.5, 200 mM NaCl, 2.0 mM TCEP, 0.05% (v/v) tween-20 filtered sterilised with 5 mM glucose-6-phosphate | ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Support film - Film thickness: 12.0 nm / Pretreatment - Type: PLASMA CLEANING | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 81000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Average exposure time: 1.22 sec. / Average electron dose: 55.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 116.12 / Target criteria: correlation coefficient | ||||||
| Output model |  PDB-7q12: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)























































