[English] 日本語
 Yorodumi
Yorodumi- EMDB-13440: Cryo-EM helical reconstruction of E. coli TnsB in complex with ri... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM helical reconstruction of E. coli TnsB in complex with right end fragment of Tn7 transposon | |||||||||
 Map data Map data | Sharpened and symmetrized | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Escherichia coli (E. coli) Escherichia coli (E. coli) | |||||||||
| Method | helical reconstruction /  cryo EM / Resolution: 3.79 Å cryo EM / Resolution: 3.79 Å | |||||||||
 Authors Authors | Czarnocki-Cieciura M / Kaczmarska Z | |||||||||
| Funding support | European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2022 Journal: Mol Cell / Year: 2022Title: Structural basis of transposon end recognition explains central features of Tn7 transposition systems. Authors: Zuzanna Kaczmarska / Mariusz Czarnocki-Cieciura / Karolina M Górecka-Minakowska / Robert J Wingo / Justyna Jackiewicz / Weronika Zajko / Jarosław T Poznański / Michał Rawski / Timothy ...Authors: Zuzanna Kaczmarska / Mariusz Czarnocki-Cieciura / Karolina M Górecka-Minakowska / Robert J Wingo / Justyna Jackiewicz / Weronika Zajko / Jarosław T Poznański / Michał Rawski / Timothy Grant / Joseph E Peters / Marcin Nowotny /   Abstract: Tn7 is a bacterial transposon with relatives containing element-encoded CRISPR-Cas systems mediating RNA-guided transposon insertion. Here, we present the 2.7 Å cryoelectron microscopy structure of ...Tn7 is a bacterial transposon with relatives containing element-encoded CRISPR-Cas systems mediating RNA-guided transposon insertion. Here, we present the 2.7 Å cryoelectron microscopy structure of prototypic Tn7 transposase TnsB interacting with the transposon end DNA. When TnsB interacts across repeating binding sites, it adopts a beads-on-a-string architecture, where the DNA-binding and catalytic domains are arranged in a tiled and intertwined fashion. The DNA-binding domains form few base-specific contacts leading to a binding preference that requires multiple weakly conserved sites at the appropriate spacing to achieve DNA sequence specificity. TnsB binding imparts differences in the global structure of the protein-bound DNA ends dictated by the spacing or overlap of binding sites explaining functional differences in the left and right ends of the element. We propose a model of the strand-transfer complex in which the terminal TnsB molecule is rearranged so that its catalytic domain is in a position conducive to transposition. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13440.map.gz emd_13440.map.gz | 104.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13440-v30.xml emd-13440-v30.xml emd-13440.xml emd-13440.xml | 22.1 KB 22.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13440_fsc.xml emd_13440_fsc.xml | 17.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_13440.png emd_13440.png | 183.7 KB | ||
| Masks |  emd_13440_msk_1.map emd_13440_msk_1.map | 512 MB |  Mask map Mask map | |
| Others |  emd_13440_additional_1.map.gz emd_13440_additional_1.map.gz emd_13440_half_map_1.map.gz emd_13440_half_map_1.map.gz emd_13440_half_map_2.map.gz emd_13440_half_map_2.map.gz | 250.5 MB 474.9 MB 474.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13440 http://ftp.pdbj.org/pub/emdb/structures/EMD-13440 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13440 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13440 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13440.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13440.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened and symmetrized | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_13440_msk_1.map emd_13440_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
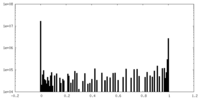
| Density Histograms |
-Additional map: Raw map
| File | emd_13440_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Raw map | ||||||||||||
| Projections & Slices |
| ||||||||||||
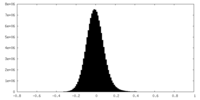
| Density Histograms |
-Half map: Half map B
| File | emd_13440_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A
| File | emd_13440_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : TnsB-DNA complex
| Entire | Name: TnsB-DNA complex |
|---|---|
| Components |
|
-Supramolecule #1: TnsB-DNA complex
| Supramolecule | Name: TnsB-DNA complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: TnsB from the canonical E. coli Tn7 element in complex with the right transposon end fragment |
|---|---|
| Molecular weight | Theoretical: 43 KDa |
-Supramolecule #2: TnsB
| Supramolecule | Name: TnsB / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 / Details: TnsB from the canonical E. coli Tn7 element |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Supramolecule #3: Tn7 transposon right end fragment
| Supramolecule | Name: Tn7 transposon right end fragment / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 / Details: E. coli Tn7 transposon right end fragment |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Macromolecule #1: Transposon Tn7 transposition protein TnsB
| Macromolecule | Name: Transposon Tn7 transposition protein TnsB / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: SMWQINEVVL FDNDPYRILA IEDGQVVWMQ ISADKGVPQA RAELLLMQYL DEGRLVRTDD P YVHLDLEE PSVDSVSFQK REEDYRKILP IINSKDRFDP KVRSELVEHV VQEHKVTKAT VY KLLRRYW QRGQTPNALI PDYKNSGAPG ERRSATGTAK IGRAREYGKG ...String: SMWQINEVVL FDNDPYRILA IEDGQVVWMQ ISADKGVPQA RAELLLMQYL DEGRLVRTDD P YVHLDLEE PSVDSVSFQK REEDYRKILP IINSKDRFDP KVRSELVEHV VQEHKVTKAT VY KLLRRYW QRGQTPNALI PDYKNSGAPG ERRSATGTAK IGRAREYGKG EGTKVTPEIE RLF RLTIEK HLLNQKGTKT TVAYRRFVDL FAQYFPRIPQ EDYPTLRQFR YFYDREYPKA QRLK SRVKA GVYKKDVRPL SSTATSQALG PGSRYEIDAT IADIYLVDHH DRQKIIGRPT LYIVI DVFS RMITGFYIGF ENPSYVVAMQ AFVNACSDKT AICAQHDIEI SSSDWPCVGL PDVLLA DRG ELMSHQVEAL VSSFNVRVES APPRRGDAKG IVESTFRTLQ AEFKSFAPGI VEGSRIK SH GETDYRLDAS LSVFEFTQII LRTILFRNNH LVMDKYDRDA DFPTDLPSIP VQLWQWGM Q HRTGSLRAVE QEQLRVALLP RRKVSISSFG VNLWGLYYSG SEILREGWLQ RSTDIARPQ HLEAAYDPVL VDTIYLFPQV GSRVFWRCNL TERSRQFKGL SFWEVWDIQA QEKHNKANAK QDELTKRRE LEAFIQQTIQ KANKLTPSTT EPKSTRIKQI KTNKKEAVTS ERKKRAEHLK P SSSGDEAK VIPFNAVEAD DQEDYSLPTY VPELFQDPPE KDES |
-Macromolecule #2: Right end fragment of Tn7 transposon chain 1
| Macromolecule | Name: Right end fragment of Tn7 transposon chain 1 / type: dna / ID: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: CTAGTTTAAG ACTTTATTGT CATAGTTTAG ATCTATTTTG TTCAGTTTAA GACTTTATTG TCCGCCCACA |
-Macromolecule #3: Right end fragment of Tn7 transposon chain 2
| Macromolecule | Name: Right end fragment of Tn7 transposon chain 2 / type: dna / ID: 3 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: TGTGGGCGGA CAATAAAGTC TTAAACTGAA CAAAATAGAT CTAAACTATG ACAATAAAGT CTTAAACTAG |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 4 s blot time, -5 blot force.. | |||||||||
| Details | Sample fixed with 0.05% glutaraldehyde and concentrated prior to vitrification; exact concentration cannot be estimated accurately. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X