[English] 日本語
 Yorodumi
Yorodumi- EMDB-13351: cryo-EM structure of DEPTOR bound to human mTOR complex 1, focuss... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13351 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
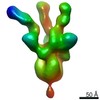
| Title | cryo-EM structure of DEPTOR bound to human mTOR complex 1, focussed on one protomer | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of TORC2 signaling / regulation of extrinsic apoptotic signaling pathway / RNA polymerase III type 2 promoter sequence-specific DNA binding / positive regulation of cytoplasmic translational initiation / RNA polymerase III type 1 promoter sequence-specific DNA binding / positive regulation of pentose-phosphate shunt / T-helper 1 cell lineage commitment / regulation of locomotor rhythm / positive regulation of wound healing, spreading of epidermal cells / cellular response to leucine starvation ...negative regulation of TORC2 signaling / regulation of extrinsic apoptotic signaling pathway / RNA polymerase III type 2 promoter sequence-specific DNA binding / positive regulation of cytoplasmic translational initiation / RNA polymerase III type 1 promoter sequence-specific DNA binding / positive regulation of pentose-phosphate shunt / T-helper 1 cell lineage commitment / regulation of locomotor rhythm / positive regulation of wound healing, spreading of epidermal cells / cellular response to leucine starvation / TFIIIC-class transcription factor complex binding /  TORC2 complex / heart valve morphogenesis / TORC2 complex / heart valve morphogenesis /  regulation of membrane permeability / negative regulation of lysosome organization / RNA polymerase III type 3 promoter sequence-specific DNA binding / regulation of membrane permeability / negative regulation of lysosome organization / RNA polymerase III type 3 promoter sequence-specific DNA binding /  TORC1 complex / positive regulation of transcription of nucleolar large rRNA by RNA polymerase I / calcineurin-NFAT signaling cascade / TORC1 complex / positive regulation of transcription of nucleolar large rRNA by RNA polymerase I / calcineurin-NFAT signaling cascade /  regulation of autophagosome assembly / TORC1 signaling / positive regulation of odontoblast differentiation / voluntary musculoskeletal movement / regulation of osteoclast differentiation / positive regulation of keratinocyte migration / cellular response to L-leucine / MTOR signalling / Amino acids regulate mTORC1 / cellular response to nutrient / energy reserve metabolic process / Energy dependent regulation of mTOR by LKB1-AMPK / regulation of autophagosome assembly / TORC1 signaling / positive regulation of odontoblast differentiation / voluntary musculoskeletal movement / regulation of osteoclast differentiation / positive regulation of keratinocyte migration / cellular response to L-leucine / MTOR signalling / Amino acids regulate mTORC1 / cellular response to nutrient / energy reserve metabolic process / Energy dependent regulation of mTOR by LKB1-AMPK /  phosphatidic acid binding / nucleus localization / negative regulation of TOR signaling / ruffle organization / protein serine/threonine kinase inhibitor activity / negative regulation of cell size / cellular response to osmotic stress / positive regulation of osteoclast differentiation / enzyme-substrate adaptor activity / phosphatidic acid binding / nucleus localization / negative regulation of TOR signaling / ruffle organization / protein serine/threonine kinase inhibitor activity / negative regulation of cell size / cellular response to osmotic stress / positive regulation of osteoclast differentiation / enzyme-substrate adaptor activity /  anoikis / cardiac muscle cell development / positive regulation of transcription by RNA polymerase III / negative regulation of protein localization to nucleus / anoikis / cardiac muscle cell development / positive regulation of transcription by RNA polymerase III / negative regulation of protein localization to nucleus /  regulation of myelination / negative regulation of calcineurin-NFAT signaling cascade / regulation of myelination / negative regulation of calcineurin-NFAT signaling cascade /  Macroautophagy / Macroautophagy /  regulation of cell size / negative regulation of macroautophagy / lysosome organization / positive regulation of oligodendrocyte differentiation / positive regulation of actin filament polymerization / protein kinase activator activity / regulation of cell size / negative regulation of macroautophagy / lysosome organization / positive regulation of oligodendrocyte differentiation / positive regulation of actin filament polymerization / protein kinase activator activity /  protein kinase inhibitor activity / positive regulation of myotube differentiation / behavioral response to pain / protein kinase inhibitor activity / positive regulation of myotube differentiation / behavioral response to pain /  TOR signaling / oligodendrocyte differentiation / mTORC1-mediated signalling / germ cell development / Constitutive Signaling by AKT1 E17K in Cancer / TOR signaling / oligodendrocyte differentiation / mTORC1-mediated signalling / germ cell development / Constitutive Signaling by AKT1 E17K in Cancer /  social behavior / cellular response to nutrient levels / CD28 dependent PI3K/Akt signaling / positive regulation of phosphoprotein phosphatase activity / positive regulation of translational initiation / neuronal action potential / HSF1-dependent transactivation / positive regulation of TOR signaling / positive regulation of G1/S transition of mitotic cell cycle / positive regulation of epithelial to mesenchymal transition / social behavior / cellular response to nutrient levels / CD28 dependent PI3K/Akt signaling / positive regulation of phosphoprotein phosphatase activity / positive regulation of translational initiation / neuronal action potential / HSF1-dependent transactivation / positive regulation of TOR signaling / positive regulation of G1/S transition of mitotic cell cycle / positive regulation of epithelial to mesenchymal transition /  regulation of macroautophagy / regulation of macroautophagy /  endomembrane system / 'de novo' pyrimidine nucleobase biosynthetic process / response to amino acid / positive regulation of lipid biosynthetic process / phagocytic vesicle / positive regulation of lamellipodium assembly / positive regulation of autophagy / heart morphogenesis / regulation of cellular response to heat / cytoskeleton organization / cardiac muscle contraction / negative regulation of TORC1 signaling / positive regulation of stress fiber assembly / cellular response to amino acid starvation / T cell costimulation / cellular response to starvation / positive regulation of endothelial cell proliferation / positive regulation of glycolytic process / protein serine/threonine kinase activator activity / response to nutrient levels / post-embryonic development / response to nutrient / negative regulation of autophagy / positive regulation of translation / 14-3-3 protein binding / VEGFR2 mediated vascular permeability / positive regulation of peptidyl-threonine phosphorylation / Regulation of PTEN gene transcription endomembrane system / 'de novo' pyrimidine nucleobase biosynthetic process / response to amino acid / positive regulation of lipid biosynthetic process / phagocytic vesicle / positive regulation of lamellipodium assembly / positive regulation of autophagy / heart morphogenesis / regulation of cellular response to heat / cytoskeleton organization / cardiac muscle contraction / negative regulation of TORC1 signaling / positive regulation of stress fiber assembly / cellular response to amino acid starvation / T cell costimulation / cellular response to starvation / positive regulation of endothelial cell proliferation / positive regulation of glycolytic process / protein serine/threonine kinase activator activity / response to nutrient levels / post-embryonic development / response to nutrient / negative regulation of autophagy / positive regulation of translation / 14-3-3 protein binding / VEGFR2 mediated vascular permeability / positive regulation of peptidyl-threonine phosphorylation / Regulation of PTEN gene transcriptionSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.67 Å cryo EM / Resolution: 3.67 Å | |||||||||
 Authors Authors | Waelchli M / Maier T | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2021 Journal: Elife / Year: 2021Title: Regulation of human mTOR complexes by DEPTOR. Authors: Matthias Wälchli / Karolin Berneiser / Francesca Mangia / Stefan Imseng / Louise-Marie Craigie / Edward Stuttfeld / Michael N Hall / Timm Maier /  Abstract: The vertebrate-specific DEP domain-containing mTOR interacting protein (DEPTOR), an oncoprotein or tumor suppressor, has important roles in metabolism, immunity, and cancer. It is the only protein ...The vertebrate-specific DEP domain-containing mTOR interacting protein (DEPTOR), an oncoprotein or tumor suppressor, has important roles in metabolism, immunity, and cancer. It is the only protein that binds and regulates both complexes of mammalian target of rapamycin (mTOR), a central regulator of cell growth. Biochemical analysis and cryo-EM reconstructions of DEPTOR bound to human mTOR complex 1 (mTORC1) and mTORC2 reveal that both structured regions of DEPTOR, the PDZ domain and the DEP domain tandem (DEPt), are involved in mTOR interaction. The PDZ domain binds tightly with mildly activating effect, but then acts as an anchor for DEPt association that allosterically suppresses mTOR activation. The binding interfaces of the PDZ domain and DEPt also support further regulation by other signaling pathways. A separate, substrate-like mode of interaction for DEPTOR phosphorylation by mTOR complexes rationalizes inhibition of non-stimulated mTOR activity at higher DEPTOR concentrations. The multifaceted interplay between DEPTOR and mTOR provides a basis for understanding the divergent roles of DEPTOR in physiology and opens new routes for targeting the mTOR-DEPTOR interaction in disease. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13351.map.gz emd_13351.map.gz | 395.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13351-v30.xml emd-13351-v30.xml emd-13351.xml emd-13351.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13351.png emd_13351.png | 98.4 KB | ||
| Masks |  emd_13351_msk_1.map emd_13351_msk_1.map | 421.9 MB |  Mask map Mask map | |
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13351 http://ftp.pdbj.org/pub/emdb/structures/EMD-13351 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13351 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13351 | HTTPS FTP |
-Related structure data
| Related structure data |  7pebMC  7pe7C  7pe8C  7pe9C  7peaC  7pecC  7pedC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13351.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13351.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.112 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13351_msk_1.map emd_13351_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : mTORC1 in complex with its regulator DEPTOR
| Entire | Name: mTORC1 in complex with its regulator DEPTOR |
|---|---|
| Components |
|
-Supramolecule #1: mTORC1 in complex with its regulator DEPTOR
| Supramolecule | Name: mTORC1 in complex with its regulator DEPTOR / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Molecular weight | Theoretical: 1.04 MDa |
-Macromolecule #1: Serine/threonine-protein kinase mTOR
| Macromolecule | Name: Serine/threonine-protein kinase mTOR / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number:  non-specific serine/threonine protein kinase non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 287.484156 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MLGTGPAAAT TAATTS(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)SRNEET(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)EMSQE ...String: MLGTGPAAAT TAATTS(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)SRNEET(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)EMSQE ESTRFYDQLN HHIFELVSSS DANERKGGIL AIASLIGVEG GNATRIGRFA NYLRNLLPS NDPVVMEMAS KAIGRLAMAG DTFTAEYVEF EVKRALEWLG ADRNEGRRHA AVLVLRELAI SVPTFFFQQV Q PFFDNIFV AVWDPKQAIR EGAVAALRAC LILTTQREPK EMQKPQWYRH TFEEAEKGFD ETLAKEKGMN RDDRIHGALL IL NELVRIS SMEGERLREE MEEITQQQLV HDKYCKDLMG FGTKPRHITP FTSFQAVQPQ QSNALVGLLG YSSHQGLMGF GTS PSPAKS T(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)RNS KNSLIQMTIL NLLPRLAAFR PS AFTDTQY LQDTMNHVLS CVKKEKERTA AFQALGLLSV AVRSEFKVYL PRVLDIIRAA LPPKDFAHKR QKAMQVDATV FTC ISMLAR AMGPGIQQDI KELLEPMLAV GLSPALTAVL YDLSRQIPQL KKDIQDGLLK MLSLVLMHKP LRHPGMPKGL AHQL ASPGL TTLPEASDVG SITLALRTLG SFEFEGHSLT QFVRHCADHF LNSEHKEIRM EAARTCSRLL TPSIHLISGH AHVVS QTAV QVVADVLSKL LVVGITDPDP DIRYCVLASL DERFDAHLAQ AENLQALFVA LNDQVFEIRE LAICTVGRLS SMNPAF VMP FLRKMLIQIL TELEHSGIGR IKEQSARMLG HLVSNAPRLI RPYMEPILKA LILKLKDPDP DPNPGVINNV LATIGEL AQ VSGLEMRKWV DELFIIIMDM LQDSSLLAKR QVALWTLGQL VASTGYVVEP YRKYPTLLEV LLNFLKTEQN QGTRREAI R VLGLLGALDP YKHKVNIGMI DQSRDASAVS LSESKSSQDS SDYSTSEMLV NMGNLPLDEF YPAVSMVALM RIFRDQSLS HHHTMVVQAI TFIFKSLGLK CVQFLPQVMP TFLNVIRVCD GAIREFLFQQ LGMLVSFVKS HIRPYMDEIV TLMREFWVMN TSIQSTIIL LIEQIVVALG GEFKLYLPQL IPHMLRVFMH DNSPGRIVSI KLLAAIQLFG ANLDDYLHLL LPPIVKLFDA P EAPLPSRK AALETVDRLT ESLDFTDYAS RIIHPIVRTL DQSPELRSTA MDTLSSLVFQ LGKKYQIFIP MVNKVLVRHR IN HQRYDVL ICRIVKGYTL ADEEEDPLIY QHRMLRSGQG DALASGPVET GPMKKLHVST INLQKAWGAA RRVSKDDWLE WLR RLSLEL LKDSSSPSLR SCWALAQAYN PMARDLFNAA FVSCWSELNE DQQDELIRSI ELALTSQDIA EVTQTLLNLA EFME HSDKG PLPLRDDNGI VLLGERAAKC RAYAKALHYK ELEFQKGPTP AILESLISIN NKLQQPEAAA GVLEYAMKHF GELEI QATW YEKLHEWEDA LVAYDKKMDT NKDDPELMLG RMRCLEALGE WGQLHQQCCE KWTLVNDETQ AKMARMAAAA AWGLGQ WDS MEEYTCMIPR DTHDGAFYRA VLALHQDLFS LAQQCIDKAR DLLDAELTAM AGESYSRAYG AMVSCHMLSE LEEVIQY KL VPERREIIRQ IWWERLQGCQ RIVEDWQKIL MVRSLVVSPH EDMRTWLKYA SLCGKSGRLA LAHKTLVLLL GVDPSRQL D HPLPTVHPQV TYAYMKNMWK SARKIDAFQH MQHFVQTMQQ QAQHAIATED QQHKQELHKL MARCFLKLGE WQLNLQGIN ESTIPKVLQY YSAATEHDRS WYKAWHAWAV MNFEAVLHYK HQNQARDEKK KLRHASGANI TNATTAATTA ATATTTASTE GSNSESEAE STENSPTPSP LQKKVTEDLS KTLLMYTVPA VQGFFRSISL SRGNNLQDTL RVLTLWFDYG HWPDVNEALV E GVKAIQID TWLQVIPQLI ARIDTPRPLV GRLIHQLLTD IGRYHPQALI YPLTVASKST TTARHNAANK ILKNMCEHSN TL VQQAMMV SEELIRVAIL WHEMWHEGLE EASRLYFGER NVKGMFEVLE PLHAMMERGP QTLKETSFNQ AYGRDLMEAQ EWC RKYMKS GNVKDLTQAW DLYYHVFRRI SKQLPQLTSL ELQYVSPKLL MCRDLELAVP GTYDPNQPII RIQSIAPSLQ VITS KQRPR KLTLMGSNGH EFVFLLKGHE DLRQDERVMQ LFGLVNTLLA NDPTSLRKNL SIQRYAVIPL STNSGLIGWV PHCDT LHAL IRDYREKKKI LLNIEHRIML RMAPDYDHLT LMQKVEVFEH AVNNTAGDDL AKLLWLKSPS SEVWFDRRTN YTRSLA VMS MVGYILGLGD RHPSNLMLDR LSGKILHIDF GDCFEVAMTR EKFPEKIPFR LTRMLTNAME VTGLDGNYRI TCHTVME VL REHKDSVMAV LEAFVYDPLL NWRLMDTNTK GNKRSRTRTD SYSAGQSVEI LDGVELGEPA HKKTGTTVPE SIHSFIGD G LVKPEALNKK AIQIINRVRD KLTGRDFSHD DTLDVPTQVE LLIKQATSHE NLCQCYIGWC PFW |
-Macromolecule #2: Target of rapamycin complex subunit LST8
| Macromolecule | Name: Target of rapamycin complex subunit LST8 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 35.91009 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MNTSPGTVGS DPVILATAGY DHTVRFWQAH SGICTRTVQH QDSQVNALEV TPDRSMIAAA GYQHIRMYDL NSNNPNPIIS YDGVNKNIA SVGFHEDGRW MYTGGEDCTA RIWDLRSRNL QCQRIFQVNA PINCVCLHPN QAELIVGDQS GAIHIWDLKT D HNEQLIPE ...String: MNTSPGTVGS DPVILATAGY DHTVRFWQAH SGICTRTVQH QDSQVNALEV TPDRSMIAAA GYQHIRMYDL NSNNPNPIIS YDGVNKNIA SVGFHEDGRW MYTGGEDCTA RIWDLRSRNL QCQRIFQVNA PINCVCLHPN QAELIVGDQS GAIHIWDLKT D HNEQLIPE PEVSITSAHI DPDASYMAAV NSTGNCYVWN LTGGIGDEVT QLIPKTKIPA HTRYALQCRF SPDSTLLATC SA DQTCKIW RTSNFSLMTE LSIKSGNPGE SSRGWMWGCA FSGDSQYIVT ASSDNLARLW CVETGEIKRE YGGHQKAVVC LAF NDSVLG |
-Macromolecule #3: Regulatory-associated protein of mTOR
| Macromolecule | Name: Regulatory-associated protein of mTOR / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 155.963094 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MAHHHHHHHH HHGSTSGSGE QKLISEEDLG STSGSGDYKD DDDKLTSLYK KAGLENLYFQ GMESEMLQSP LLGLGEEDEA DLTDWNLPL AFMKKRHCEK IEGSKSLAQS WRMKDRMKTV SVALVLCLNV GVDPPDVVKT TPCARLECWI DPLSMGPQKA L ETIGANLQ ...String: MAHHHHHHHH HHGSTSGSGE QKLISEEDLG STSGSGDYKD DDDKLTSLYK KAGLENLYFQ GMESEMLQSP LLGLGEEDEA DLTDWNLPL AFMKKRHCEK IEGSKSLAQS WRMKDRMKTV SVALVLCLNV GVDPPDVVKT TPCARLECWI DPLSMGPQKA L ETIGANLQ KQYENWQPRA RYKQSLDPTV DEVKKLCTSL RRNAKEERVL FHYNGHGVPR PTVNGEVWVF NKNYTQYIPL SI YDLQTWM GSPSIFVYDC SNAGLIVKSF KQFALQREQE LEVAAINPNH PLAQMPLPPS MKNCIQLAAC EATELLPMIP DLP ADLFTS CLTTPIKIAL RWFCMQKCVS LVPGVTLDLI EKIPGRLNDR RTPLGELNWI FTAITDTIAW NVLPRDLFQK LFRQ DLLVA SLFRNFLLAE RIMRSYNCTP VSSPRLPPTY MHAMWQAWDL AVDICLSQLP TIIEEGTAFR HSPFFAEQLT AFQVW LTMG VENRNPPEQL PIVLQVLLSQ VHRLRALDLL GRFLDLGPWA VSLALSVGIF PYVLKLLQSS ARELRPLLVF IWAKIL AVD SSCQADLVKD NGHKYFLSVL ADPYMPAEHR TMTAFILAVI VNSYHTGQEA CLQGNLIAIC LEQLNDPHPL LRQWVAI CL GRIWQNFDSA RWCGVRDSAH EKLYSLLSDP IPEVRCAAVF ALGTFVGNSA ERTDHSTTID HNVAMMLAQL VSDGSPMV R KELVVALSHL VVQYESNFCT VALQFIEEEK NYALPSPATT EGGSLTPVRD SPCTPRLRSV SSYGNIRAVA TARSLNKSL QNLSLTEESG GAVAFSPGNL STSSSASSTL GSPENEEHIL SFETIDKMRR ASSYSSLNSL IGVSFNSVYT QIWRVLLHLA ADPYPEVSD VAMKVLNSIA YKATVNARPQ RVLDTSSLTQ SAPASPTNKG VHIHQAGGSP PASSTSSSSL TNDVAKQPVS R DLPSGRPG TTGPAGAQYT PHSHQFPRTR KMFDKGPEQT ADDADDAAGH KSFISATVQT GFCDWSARYF AQPVMKIPEE HD LESQIRK EREWRFLRNS RVRRQAQQVI QKGITRLDDQ IFLNRNPGVP SVVKFHPFTP CIAVADKDSI CFWDWEKGEK LDY FHNGNP RYTRVTAMEY LNGQDCSLLL TATDDGAIRV WKNFADLEKN PEMVTAWQGL SDMLPTTRGA GMVVDWEQET GLLM SSGDV RIVRIWDTDR EMKVQDIPTG ADSCVTSLSC DSHRSLIVAG LGDGSIRVYD RRMALSECRV MTYREHTAWV VKASL QKRP DGHIVSVSVN GDVRIFDPRM PESVNVLQIV KGLTALDIHP QADLIACGSV NQFTAIYNSS GELINNIKYY DGFMGQ RVG AISCLAFHPH WPHLAVGSND YYISVYSVEK RVR |
-Macromolecule #4: DEP domain-containing mTOR-interacting protein
| Macromolecule | Name: DEP domain-containing mTOR-interacting protein / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 46.365832 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MEEGGSTGSA GSDSSTSGSG GAQQRELERM AEVLVTGEQL RLRLHEEKVI KDRRHHLKTY PNCFVAKELI DWLIEHKEAS DRETAIKLM QKLADRGIIH HVCDEHKEFK DVKLFYRFRK DDGTFPLDNE VKAFMRGQRL YEKLMSPENT LLQPREEEGV K YERTFMAS ...String: MEEGGSTGSA GSDSSTSGSG GAQQRELERM AEVLVTGEQL RLRLHEEKVI KDRRHHLKTY PNCFVAKELI DWLIEHKEAS DRETAIKLM QKLADRGIIH HVCDEHKEFK DVKLFYRFRK DDGTFPLDNE VKAFMRGQRL YEKLMSPENT LLQPREEEGV K YERTFMAS EFLDWLVQEG EATTRKEAEQ LCHRLMEHGI IQHVSSKHPF VDSNLLYQFR MNFRRRRRLM ELLNEKSPSS QE THDSPFC LRKQSHDNRK STSFMSVSPS KEIKIVSAVR RSSMSSCGSS GYFSSSPTLS SSPPVLCNPK SVLKRPVTSE ELL TPGAPY ARKTFTIVGD AVGWGFVVRG SKPCHIQAVD PSGPAAAAGM KVCQFVVSVN GLNVLHVDYR TVNNLILTGP RTIV MEVME ELEC |
-Macromolecule #5: INOSITOL HEXAKISPHOSPHATE
| Macromolecule | Name: INOSITOL HEXAKISPHOSPHATE / type: ligand / ID: 5 / Number of copies: 1 / Formula: IHP |
|---|---|
| Molecular weight | Theoretical: 660.035 Da |
| Chemical component information |  ChemComp-IHP: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.4 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
- Image processing
Image processing
| CTF correction | Software - Name: cryoSPARC (ver. 3.2.0) |
|---|---|
| Initial angle assignment | Type: RANDOM ASSIGNMENT / Software - Name: cryoSPARC (ver. 3.2.0) |
| Final 3D classification | Number classes: 3 / Software - Name: cryoSPARC (ver. 3.2.0) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 3.2.0) |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.67 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 3.2.0) / Number images used: 850152 |
 Movie
Movie Controller
Controller




























 Z
Z Y
Y X
X










