[English] 日本語
 Yorodumi
Yorodumi- EMDB-12978: structure of human haemoglobin obtained via cryoelectron microsco... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12978 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | structure of human haemoglobin obtained via cryoelectron microscopy at 200 kV | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationnitric oxide transport / hemoglobin alpha binding / haptoglobin-hemoglobin complex /  organic acid binding / cellular oxidant detoxification / organic acid binding / cellular oxidant detoxification /  hemoglobin binding / renal absorption / hemoglobin binding / renal absorption /  hemoglobin complex / hemoglobin complex /  oxygen transport / Scavenging of heme from plasma ...nitric oxide transport / hemoglobin alpha binding / haptoglobin-hemoglobin complex / oxygen transport / Scavenging of heme from plasma ...nitric oxide transport / hemoglobin alpha binding / haptoglobin-hemoglobin complex /  organic acid binding / cellular oxidant detoxification / organic acid binding / cellular oxidant detoxification /  hemoglobin binding / renal absorption / hemoglobin binding / renal absorption /  hemoglobin complex / hemoglobin complex /  oxygen transport / Scavenging of heme from plasma / endocytic vesicle lumen / blood vessel diameter maintenance / hydrogen peroxide catabolic process / oxygen transport / Scavenging of heme from plasma / endocytic vesicle lumen / blood vessel diameter maintenance / hydrogen peroxide catabolic process /  oxygen carrier activity / Late endosomal microautophagy / Heme signaling / carbon dioxide transport / response to hydrogen peroxide / Erythrocytes take up oxygen and release carbon dioxide / Erythrocytes take up carbon dioxide and release oxygen / Cytoprotection by HMOX1 / oxygen carrier activity / Late endosomal microautophagy / Heme signaling / carbon dioxide transport / response to hydrogen peroxide / Erythrocytes take up oxygen and release carbon dioxide / Erythrocytes take up carbon dioxide and release oxygen / Cytoprotection by HMOX1 /  platelet aggregation / platelet aggregation /  oxygen binding / oxygen binding /  regulation of blood pressure / Chaperone Mediated Autophagy / positive regulation of nitric oxide biosynthetic process / tertiary granule lumen / Factors involved in megakaryocyte development and platelet production / blood microparticle / ficolin-1-rich granule lumen / iron ion binding / regulation of blood pressure / Chaperone Mediated Autophagy / positive regulation of nitric oxide biosynthetic process / tertiary granule lumen / Factors involved in megakaryocyte development and platelet production / blood microparticle / ficolin-1-rich granule lumen / iron ion binding /  heme binding / Neutrophil degranulation / heme binding / Neutrophil degranulation /  extracellular space / extracellular exosome / extracellular region / extracellular space / extracellular exosome / extracellular region /  membrane / membrane /  metal ion binding / metal ion binding /  cytosol cytosolSimilarity search - Function | |||||||||
| Biological species |   Bos taurus (cattle) Bos taurus (cattle) | |||||||||
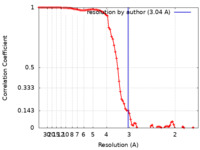
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.04 Å cryo EM / Resolution: 3.04 Å | |||||||||
 Authors Authors | Mann D / Sachse C | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: structure of human haemoglobin obtained via cryoelectron microscopy at 200 kV Authors: Mann D / Sachse C | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12978.map.gz emd_12978.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12978-v30.xml emd-12978-v30.xml emd-12978.xml emd-12978.xml | 14.8 KB 14.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12978_fsc.xml emd_12978_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_12978.png emd_12978.png | 68.3 KB | ||
| Masks |  emd_12978_msk_1.map emd_12978_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_12978_half_map_1.map.gz emd_12978_half_map_1.map.gz emd_12978_half_map_2.map.gz emd_12978_half_map_2.map.gz | 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12978 http://ftp.pdbj.org/pub/emdb/structures/EMD-12978 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12978 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12978 | HTTPS FTP |
-Related structure data
| Similar structure data | |
|---|---|
| EM raw data |  EMPIAR-10721 (Title: CryoEM single particle analysis of 65 kDa human haemoglobin using a 200 kV Talos Arctica EMPIAR-10721 (Title: CryoEM single particle analysis of 65 kDa human haemoglobin using a 200 kV Talos ArcticaData size: 1.5 TB Data #1: Unaligned movies (tiff) and aligned micrographs (mrc) of hHGB in vitreous ice [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12978.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12978.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.8389 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12978_msk_1.map emd_12978_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
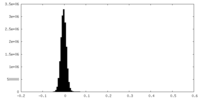
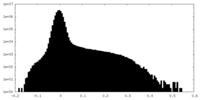
| Density Histograms |
-Half map: #1
| File | emd_12978_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_12978_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
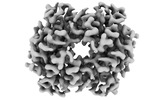
-Entire : tetrameric complex of human haemoglobin alpha and beta subunits
| Entire | Name: tetrameric complex of human haemoglobin alpha and beta subunits |
|---|---|
| Components |
|
-Supramolecule #1: tetrameric complex of human haemoglobin alpha and beta subunits
| Supramolecule | Name: tetrameric complex of human haemoglobin alpha and beta subunits type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Bos taurus (cattle) Bos taurus (cattle) |
-Macromolecule #1: haemoglobin subunit alpha
| Macromolecule | Name: haemoglobin subunit alpha / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Bos taurus (cattle) Bos taurus (cattle) |
| Sequence | String: VLSPADKTNV KAAWGKVGAH AGEYGAEALE RMFLSFPTTK TYFPHFDLSH GSAQVKGHGK KVADALTNAV AHVDDMPNAL SALSDLHAHK LRVDPVNFKL LSHCLLVTLA AHLPAEFTPA VHASLDKFLA SVSTVLTSKY R |
-Macromolecule #2: haemoglobin subunit beta
| Macromolecule | Name: haemoglobin subunit beta / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Bos taurus (cattle) Bos taurus (cattle) |
| Sequence | String: VHLTPEEKSA VTALWGKVNV DEVGGEALGR LLVVYPWTQR FFESFGDLST PDAVMGNPKV KAHGKKVLGA FSDGLAHLDN LKGTFATLSE LHCDKLHVDP ENFRLLGNVL VCVLAHHFGK EFTPPVQAAY QKVVAGVANA LAHKYH |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 7.6 / Component - Formula: TRIS / Component - Name: TRIS buffer / Component - Name: TRIS buffer |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 4 K / Instrument: FEI VITROBOT MARK IV |
| Details | obtained lyophilized from commercial source (Sigma-Aldrich), dissolved in 50 mM TRIS buffer |
- Electron microscopy
Electron microscopy
| Microscope | TFS TALOS |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 100000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 100000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 1400 / Average electron dose: 65.0 e/Å2 |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|
 Movie
Movie Controller
Controller

























 Z
Z Y
Y X
X