[English] 日本語
 Yorodumi
Yorodumi- EMDB-12048: Focused map- CyclinA-CDK2-class. Ubiquitin ligation to F-box prot... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12048 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Focused map- CyclinA-CDK2-class. Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: CUL1-RBX1-SKP1-SKP2-CKSHS1-Cyclin A-CDK2-p27. Transition State 1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationcyclin-dependent protein kinase activating kinase regulator activity / regulation of lens fiber cell differentiation / negative regulation of cardiac muscle tissue regeneration / negative regulation of cyclin-dependent protein kinase activity / autophagic cell death / positive regulation of protein polyubiquitination / Parkin-FBXW7-Cul1 ubiquitin ligase complex / negative regulation of epithelial cell proliferation involved in prostate gland development / FOXO-mediated transcription of cell cycle genes /  F-box domain binding ...cyclin-dependent protein kinase activating kinase regulator activity / regulation of lens fiber cell differentiation / negative regulation of cardiac muscle tissue regeneration / negative regulation of cyclin-dependent protein kinase activity / autophagic cell death / positive regulation of protein polyubiquitination / Parkin-FBXW7-Cul1 ubiquitin ligase complex / negative regulation of epithelial cell proliferation involved in prostate gland development / FOXO-mediated transcription of cell cycle genes / F-box domain binding ...cyclin-dependent protein kinase activating kinase regulator activity / regulation of lens fiber cell differentiation / negative regulation of cardiac muscle tissue regeneration / negative regulation of cyclin-dependent protein kinase activity / autophagic cell death / positive regulation of protein polyubiquitination / Parkin-FBXW7-Cul1 ubiquitin ligase complex / negative regulation of epithelial cell proliferation involved in prostate gland development / FOXO-mediated transcription of cell cycle genes /  F-box domain binding / Phosphorylation of proteins involved in the G2/M transition by Cyclin A:Cdc2 complexes / cyclin A2-CDK1 complex / Aberrant regulation of mitotic exit in cancer due to RB1 defects / PcG protein complex / cell cycle G1/S phase transition / cellular response to luteinizing hormone stimulus / regulation of cell cycle G1/S phase transition / regulation of exit from mitosis / epithelial cell proliferation involved in prostate gland development / cullin-RING ubiquitin ligase complex / negative regulation of phosphorylation / mitotic cell cycle phase transition / negative regulation of epithelial cell apoptotic process / negative regulation of cyclin-dependent protein serine/threonine kinase activity / ubiquitin ligase activator activity / Cul7-RING ubiquitin ligase complex / Transcription of E2F targets under negative control by p107 (RBL1) and p130 (RBL2) in complex with HDAC1 / positive regulation of ubiquitin protein ligase activity / maintenance of protein location in nucleus / cyclin-dependent protein serine/threonine kinase inhibitor activity / Loss of Function of FBXW7 in Cancer and NOTCH1 Signaling / cellular response to leptin stimulus / RHO GTPases activate CIT / F-box domain binding / Phosphorylation of proteins involved in the G2/M transition by Cyclin A:Cdc2 complexes / cyclin A2-CDK1 complex / Aberrant regulation of mitotic exit in cancer due to RB1 defects / PcG protein complex / cell cycle G1/S phase transition / cellular response to luteinizing hormone stimulus / regulation of cell cycle G1/S phase transition / regulation of exit from mitosis / epithelial cell proliferation involved in prostate gland development / cullin-RING ubiquitin ligase complex / negative regulation of phosphorylation / mitotic cell cycle phase transition / negative regulation of epithelial cell apoptotic process / negative regulation of cyclin-dependent protein serine/threonine kinase activity / ubiquitin ligase activator activity / Cul7-RING ubiquitin ligase complex / Transcription of E2F targets under negative control by p107 (RBL1) and p130 (RBL2) in complex with HDAC1 / positive regulation of ubiquitin protein ligase activity / maintenance of protein location in nucleus / cyclin-dependent protein serine/threonine kinase inhibitor activity / Loss of Function of FBXW7 in Cancer and NOTCH1 Signaling / cellular response to leptin stimulus / RHO GTPases activate CIT /  male pronucleus / male pronucleus /  female pronucleus / cyclin-dependent protein serine/threonine kinase activator activity / female pronucleus / cyclin-dependent protein serine/threonine kinase activator activity /  nuclear export / cellular response to cocaine / response to glucagon / SCF-dependent proteasomal ubiquitin-dependent protein catabolic process / AKT phosphorylates targets in the cytosol / cyclin-dependent protein serine/threonine kinase regulator activity / cellular response to insulin-like growth factor stimulus / Cul4A-RING E3 ubiquitin ligase complex / nuclear export / cellular response to cocaine / response to glucagon / SCF-dependent proteasomal ubiquitin-dependent protein catabolic process / AKT phosphorylates targets in the cytosol / cyclin-dependent protein serine/threonine kinase regulator activity / cellular response to insulin-like growth factor stimulus / Cul4A-RING E3 ubiquitin ligase complex /  SCF ubiquitin ligase complex / epithelial cell apoptotic process / cellular response to antibiotic / positive regulation of intracellular estrogen receptor signaling pathway / negative regulation of kinase activity / ubiquitin ligase complex scaffold activity / positive regulation of DNA biosynthetic process / cochlea development / molecular function inhibitor activity / SCF ubiquitin ligase complex / epithelial cell apoptotic process / cellular response to antibiotic / positive regulation of intracellular estrogen receptor signaling pathway / negative regulation of kinase activity / ubiquitin ligase complex scaffold activity / positive regulation of DNA biosynthetic process / cochlea development / molecular function inhibitor activity /  protein kinase inhibitor activity / cyclin A1-CDK2 complex / cyclin E2-CDK2 complex / cyclin E1-CDK2 complex / cellular response to platelet-derived growth factor stimulus / cyclin A2-CDK2 complex / positive regulation of DNA-templated DNA replication initiation / protein kinase inhibitor activity / cyclin A1-CDK2 complex / cyclin E2-CDK2 complex / cyclin E1-CDK2 complex / cellular response to platelet-derived growth factor stimulus / cyclin A2-CDK2 complex / positive regulation of DNA-templated DNA replication initiation /  G2 Phase / cyclin-dependent protein kinase activity / cellular response to lithium ion / G2 Phase / cyclin-dependent protein kinase activity / cellular response to lithium ion /  Y chromosome / Phosphorylation of proteins involved in G1/S transition by active Cyclin E:Cdk2 complexes / positive regulation of heterochromatin formation / p53-Dependent G1 DNA Damage Response / Prolactin receptor signaling / Y chromosome / Phosphorylation of proteins involved in G1/S transition by active Cyclin E:Cdk2 complexes / positive regulation of heterochromatin formation / p53-Dependent G1 DNA Damage Response / Prolactin receptor signaling /  X chromosome / PTK6 Regulates Cell Cycle / protein monoubiquitination / regulation of cyclin-dependent protein serine/threonine kinase activity / regulation of G1/S transition of mitotic cell cycle / Constitutive Signaling by AKT1 E17K in Cancer / regulation of anaphase-promoting complex-dependent catabolic process / cullin family protein binding / Defective binding of RB1 mutants to E2F1,(E2F2, E2F3) / inner ear development / negative regulation of vascular associated smooth muscle cell proliferation / X chromosome / PTK6 Regulates Cell Cycle / protein monoubiquitination / regulation of cyclin-dependent protein serine/threonine kinase activity / regulation of G1/S transition of mitotic cell cycle / Constitutive Signaling by AKT1 E17K in Cancer / regulation of anaphase-promoting complex-dependent catabolic process / cullin family protein binding / Defective binding of RB1 mutants to E2F1,(E2F2, E2F3) / inner ear development / negative regulation of vascular associated smooth muscle cell proliferation /  regulation of DNA replication / centriole replication / Regulation of APC/C activators between G1/S and early anaphase / centrosome duplication / ubiquitin-like ligase-substrate adaptor activity / Telomere Extension By Telomerase / G0 and Early G1 / negative regulation of mitotic cell cycle / cellular response to organic cyclic compound / Activation of the pre-replicative complex / Estrogen-dependent nuclear events downstream of ESR-membrane signaling / cyclin-dependent protein kinase holoenzyme complex / protein K48-linked ubiquitination / cellular response to nitric oxide / response to amino acid / regulation of DNA replication / centriole replication / Regulation of APC/C activators between G1/S and early anaphase / centrosome duplication / ubiquitin-like ligase-substrate adaptor activity / Telomere Extension By Telomerase / G0 and Early G1 / negative regulation of mitotic cell cycle / cellular response to organic cyclic compound / Activation of the pre-replicative complex / Estrogen-dependent nuclear events downstream of ESR-membrane signaling / cyclin-dependent protein kinase holoenzyme complex / protein K48-linked ubiquitination / cellular response to nitric oxide / response to amino acid /  cyclin-dependent kinase / cyclin-dependent kinase /  Cajal body / animal organ regeneration / cyclin-dependent protein serine/threonine kinase activity / localization / Nuclear events stimulated by ALK signaling in cancer Cajal body / animal organ regeneration / cyclin-dependent protein serine/threonine kinase activity / localization / Nuclear events stimulated by ALK signaling in cancerSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
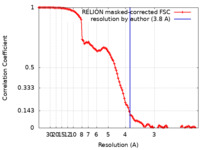
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.8 Å cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Horn-Ghetko D / Prabu JR / Schulman BA | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly. Authors: Daniel Horn-Ghetko / David T Krist / J Rajan Prabu / Kheewoong Baek / Monique P C Mulder / Maren Klügel / Daniel C Scott / Huib Ovaa / Gary Kleiger / Brenda A Schulman /    Abstract: E3 ligases are typically classified by hallmark domains such as RING and RBR, which are thought to specify unique catalytic mechanisms of ubiquitin transfer to recruited substrates. However, rather ...E3 ligases are typically classified by hallmark domains such as RING and RBR, which are thought to specify unique catalytic mechanisms of ubiquitin transfer to recruited substrates. However, rather than functioning individually, many neddylated cullin-RING E3 ligases (CRLs) and RBR-type E3 ligases in the ARIH family-which together account for nearly half of all ubiquitin ligases in humans-form E3-E3 super-assemblies. Here, by studying CRLs in the SKP1-CUL1-F-box (SCF) family, we show how neddylated SCF ligases and ARIH1 (an RBR-type E3 ligase) co-evolved to ubiquitylate diverse substrates presented on various F-box proteins. We developed activity-based chemical probes that enabled cryo-electron microscopy visualization of steps in E3-E3 ubiquitylation, initiating with ubiquitin linked to the E2 enzyme UBE2L3, then transferred to the catalytic cysteine of ARIH1, and culminating in ubiquitin linkage to a substrate bound to the SCF E3 ligase. The E3-E3 mechanism places the ubiquitin-linked active site of ARIH1 adjacent to substrates bound to F-box proteins (for example, substrates with folded structures or limited length) that are incompatible with previously described conventional RING E3-only mechanisms. The versatile E3-E3 super-assembly may therefore underlie widespread ubiquitylation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12048.map.gz emd_12048.map.gz | 8.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12048-v30.xml emd-12048-v30.xml emd-12048.xml emd-12048.xml | 23.5 KB 23.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12048_fsc.xml emd_12048_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_12048.png emd_12048.png | 54.3 KB | ||
| Masks |  emd_12048_msk_1.map emd_12048_msk_1.map | 125 MB |  Mask map Mask map | |
| Others |  emd_12048_half_map_1.map.gz emd_12048_half_map_1.map.gz emd_12048_half_map_2.map.gz emd_12048_half_map_2.map.gz | 98.4 MB 98.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12048 http://ftp.pdbj.org/pub/emdb/structures/EMD-12048 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12048 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12048 | HTTPS FTP |
-Related structure data
| Related structure data |  7b5rMC  7b5lC  7b5mC  7b5nC  7b5sC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12048.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12048.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12048_msk_1.map emd_12048_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_12048_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_12048_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : NEDD8-CUL1-RBX1-SKP1-SKP2-CKSHS1-Cyclin A-CDK2-p27-UBE2L3~Ub~ARIH...
| Entire | Name: NEDD8-CUL1-RBX1-SKP1-SKP2-CKSHS1-Cyclin A-CDK2-p27-UBE2L3~Ub~ARIH1. Transition State 1 composite map. |
|---|---|
| Components |
|
-Supramolecule #1: NEDD8-CUL1-RBX1-SKP1-SKP2-CKSHS1-Cyclin A-CDK2-p27-UBE2L3~Ub~ARIH...
| Supramolecule | Name: NEDD8-CUL1-RBX1-SKP1-SKP2-CKSHS1-Cyclin A-CDK2-p27-UBE2L3~Ub~ARIH1. Transition State 1 composite map. type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 300 KDa |
-Macromolecule #1: Cullin-1
| Macromolecule | Name: Cullin-1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 89.800367 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MSSTRSQNPH GLKQIGLDQI WDDLRAGIQQ VYTRQSMAKS RYMELYTHVY NYCTSVHQSN QARGAGVPPS KSKKGQTPGG AQFVGLELY KRLKEFLKNY LTNLLKDGED LMDESVLKFY TQQWEDYRFS SKVLNGICAY LNRHWVRREC DEGRKGIYEI Y SLALVTWR ...String: MSSTRSQNPH GLKQIGLDQI WDDLRAGIQQ VYTRQSMAKS RYMELYTHVY NYCTSVHQSN QARGAGVPPS KSKKGQTPGG AQFVGLELY KRLKEFLKNY LTNLLKDGED LMDESVLKFY TQQWEDYRFS SKVLNGICAY LNRHWVRREC DEGRKGIYEI Y SLALVTWR DCLFRPLNKQ VTNAVLKLIE KERNGETINT RLISGVVQSY VELGLNEDDA FAKGPTLTVY KESFESQFLA DT ERFYTRE STEFLQQNPV TEYMKKAEAR LLEEQRRVQV YLHESTQDEL ARKCEQVLIE KHLEIFHTEF QNLLDADKNE DLG RMYNLV SRIQDGLGEL KKLLETHIHN QGLAAIEKCG EAALNDPKMY VQTVLDVHKK YNALVMSAFN NDAGFVAALD KACG RFINN NAVTKMAQSS SKSPELLARY CDSLLKKSSK NPEEAELEDT LNQVMVVFKY IEDKDVFQKF YAKMLAKRLV HQNSA SDDA EASMISKLKQ ACGFEYTSKL QRMFQDIGVS KDLNEQFKKH LTNSEPLDLD FSIQVLSSGS WPFQQSCTFA LPSELE RSY QRFTAFYASR HSGRKLTWLY QLSKGELVTN CFKNRYTLQA STFQMAILLQ YNTEDAYTVQ QLTDSTQIKM DILAQVL QI LLKSKLLVLE DENANVDEVE LKPDTLIKLY LGYKNKKLRV NINVPMKTEQ KQEQETTHKN IEEDRKLLIQ AAIVRIMK M RKVLKHQQLL GEVLTQLSSR FKPRVPVIKK CIDILIEKEY LERVDGEKDT YSYLA |
-Macromolecule #2: S-phase kinase-associated protein 2
| Macromolecule | Name: S-phase kinase-associated protein 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 47.817785 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MHRKHLQEIP DLSSNVATSF TWGWDSSKTS ELLSGMGVSA LEKEEPDSEN IPQELLSNLG HPESPPRKRL KSKGSDKDFV IVRRPKLNR ENFPGVSWDS LPDELLLGIF SCLCLPELLK VSGVCKRWYR LASDESLWQT LDLTGKNLHP DVTGRLLSQG V IAFRCPRS ...String: MHRKHLQEIP DLSSNVATSF TWGWDSSKTS ELLSGMGVSA LEKEEPDSEN IPQELLSNLG HPESPPRKRL KSKGSDKDFV IVRRPKLNR ENFPGVSWDS LPDELLLGIF SCLCLPELLK VSGVCKRWYR LASDESLWQT LDLTGKNLHP DVTGRLLSQG V IAFRCPRS FMDQPLAEHF SPFRVQHMDL SNSVIEVSTL HGILSQCSKL QNLSLEGLRL SDPIVNTLAK NSNLVRLNLS GC SGFSEFA LQTLLSSCSR LDELNLSWCF DFTEKHVQVA VAHVSETITQ LNLSGYRKNL QKSDLSTLVR RCPNLVHLDL SDS VMLKND CFQEFFQLNY LQHLSLSRCY DIIPETLLEL GEIPTLKTLQ VFGIVPDGTL QLLKEALPHL QINCSHFTTI ARPT IGNKK NQEIWGIKCR LTLQKPSCL |
-Macromolecule #3: Cyclin-dependent kinases regulatory subunit 1
| Macromolecule | Name: Cyclin-dependent kinases regulatory subunit 1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 9.679211 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MSHKQIYYSD KYDDEEFEYR HVMLPKDIAK LVPKTHLMSE SEWRNLGVQQ SQGWVHYMIH EPEPHILLFR RPLPKKPKK |
-Macromolecule #4: S-phase kinase-associated protein 1
| Macromolecule | Name: S-phase kinase-associated protein 1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 18.679965 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MPSIKLQSSD GEIFEVDVEI AKQSVTIKTM LEDLGMDDEG DDDPVPLPNV NAAILKKVIQ WCTHHKDDPP PPEDDENKEK RTDDIPVWD QEFLKVDQGT LFELILAANY LDIKGLLDVT CKTVANMIKG KTPEEIRKTF NIKNDFTEEE EAQVRKENQW C EEK |
-Macromolecule #5: Cyclin-dependent kinase 2
| Macromolecule | Name: Cyclin-dependent kinase 2 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO / EC number:  cyclin-dependent kinase cyclin-dependent kinase |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 34.056469 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MENFQKVEKI GEGTYGVVYK ARNKLTGEVV ALKKIRLDTE TEGVPSTAIR EISLLKELNH PNIVKLLDVI HTENKLYLVF EFLHQDLKK FMDASALTGI PLPLIKSYLF QLLQGLAFCH SHRVLHRDLK PQNLLINTEG AIKLADFGLA RAFGVPVRTY (TPO)HEVVTLWY ...String: MENFQKVEKI GEGTYGVVYK ARNKLTGEVV ALKKIRLDTE TEGVPSTAIR EISLLKELNH PNIVKLLDVI HTENKLYLVF EFLHQDLKK FMDASALTGI PLPLIKSYLF QLLQGLAFCH SHRVLHRDLK PQNLLINTEG AIKLADFGLA RAFGVPVRTY (TPO)HEVVTLWY RAPEILLGCK YYSTAVDIWS LGCIFAEMVT RRALFPGDSE IDQLFRIFRT LGTPDEVVWP GVTSMPD YK PSFPKWARQD FSKVVPPLDE DGRSLLSQML HYDPNKRISA KAALAHPFFQ DVTKPVPHLR L |
-Macromolecule #6: Cyclin-A2
| Macromolecule | Name: Cyclin-A2 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 48.609574 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MLGNSAPGPA TREAGSALLA LQQTALQEDQ ENINPEKAAP VQQPRTRAAL AVLKSGNPRG LAQQQRPKTR RVAPLKDLPV NDEHVTVPP WKANSKQPAF TIHVDEAEKE AQKKPAESQK IEREDALAFN SAISLPGPRK PLVPLDYPMD GSFESPHTMD M SIILEDEK ...String: MLGNSAPGPA TREAGSALLA LQQTALQEDQ ENINPEKAAP VQQPRTRAAL AVLKSGNPRG LAQQQRPKTR RVAPLKDLPV NDEHVTVPP WKANSKQPAF TIHVDEAEKE AQKKPAESQK IEREDALAFN SAISLPGPRK PLVPLDYPMD GSFESPHTMD M SIILEDEK PVSVNEVPDY HEDIHTYLRE MEVKCKPKVG YMKKQPDITN SMRAILVDWL VEVGEEYKLQ NETLHLAVNY ID RFLSSMS VLRGKLQLVG TAAMLLASKF EEIYPPEVAE FVYITDDTYT KKQVLRMEHL VLKVLTFDLA APTVNQFLTQ YFL HQQPAN CKVESLAMFL GELSLIDADP YLKYLPSVIA GAAFHLALYT VTGQSWPESL IRKTGYTLES LKPCLMDLHQ TYLK APQHA QQSIREKYKN SKYHGVSLLN PPETLNL |
-Macromolecule #7: Cyclin-dependent kinase inhibitor 1B
| Macromolecule | Name: Cyclin-dependent kinase inhibitor 1B / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.188303 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MSNVRVSNGS PSLERMDARQ AEHPKPSACR NLFGPVDHEE LTRDLEKHCR DMEEASQRKW NFDFQNHKPL EGKYEWQEVE KGSLPEFYY RPPRPPKGAC KVPAQESQDV SGSRPAAPLI GAPANSEDTH LVDPKTDPSD SQTGLAEQCA GIRKRPATDD S STQNKRAN ...String: MSNVRVSNGS PSLERMDARQ AEHPKPSACR NLFGPVDHEE LTRDLEKHCR DMEEASQRKW NFDFQNHKPL EGKYEWQEVE KGSLPEFYY RPPRPPKGAC KVPAQESQDV SGSRPAAPLI GAPANSEDTH LVDPKTDPSD SQTGLAEQCA GIRKRPATDD S STQNKRAN RTEENVSDGS PNAGSVEQ(TPO)P KKPGLRRRQT |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 70.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












































 Z
Z Y
Y X
X