[English] 日本語
 Yorodumi
Yorodumi- EMDB-11406: Native putative AMPAR-type glutamate receptor, de novo 3D classif... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11406 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Native putative AMPAR-type glutamate receptor, de novo 3D classification | |||||||||
 Map data Map data | Averaged density map. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Rattus norvegicus (Norway rat) Rattus norvegicus (Norway rat) | |||||||||
| Method | subtomogram averaging /  cryo EM / Resolution: 31.268572 Å cryo EM / Resolution: 31.268572 Å | |||||||||
 Authors Authors | Martinez A / Lucic V | |||||||||
| Funding support | European Union,  Spain, 2 items Spain, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2021 Journal: Sci Adv / Year: 2021Title: Trans-synaptic assemblies link synaptic vesicles and neuroreceptors. Authors: Antonio Martinez-Sanchez / Ulrike Laugks / Zdravko Kochovski / Christos Papantoniou / Luca Zinzula / Wolfgang Baumeister / Vladan Lučić /   Abstract: Synaptic transmission is characterized by fast, tightly coupled processes and complex signaling pathways that require a precise protein organization, such as the previously reported nanodomain ...Synaptic transmission is characterized by fast, tightly coupled processes and complex signaling pathways that require a precise protein organization, such as the previously reported nanodomain colocalization of pre- and postsynaptic proteins. Here, we used cryo-electron tomography to visualize synaptic complexes together with their native environment comprising interacting proteins and lipids on a 2- to 4-nm scale. Using template-free detection and classification, we showed that tripartite trans-synaptic assemblies (subcolumns) link synaptic vesicles to postsynaptic receptors and established that a particular displacement between directly interacting complexes characterizes subcolumns. Furthermore, we obtained de novo average structures of ionotropic glutamate receptors in their physiological composition, embedded in plasma membrane. These data support the hypothesis that synaptic function is carried by precisely organized trans-synaptic units. It provides a framework for further exploration of synaptic and other large molecular assemblies that link different cells or cellular regions and may require weak or transient interactions to exert their function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11406.map.gz emd_11406.map.gz | 963.1 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11406-v30.xml emd-11406-v30.xml emd-11406.xml emd-11406.xml | 11.8 KB 11.8 KB | Display Display |  EMDB header EMDB header |
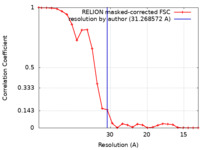
| FSC (resolution estimation) |  emd_11406_fsc.xml emd_11406_fsc.xml | 2.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_11406.png emd_11406.png | 32.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11406 http://ftp.pdbj.org/pub/emdb/structures/EMD-11406 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11406 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11406 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11406.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11406.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Averaged density map. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 6.84 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Native putative AMPA-type ionotropic glutamate receptor imaged at...
| Entire | Name: Native putative AMPA-type ionotropic glutamate receptor imaged at excitatory neocortical rodent synapses |
|---|---|
| Components |
|
-Supramolecule #1: Native putative AMPA-type ionotropic glutamate receptor imaged at...
| Supramolecule | Name: Native putative AMPA-type ionotropic glutamate receptor imaged at excitatory neocortical rodent synapses type: complex / ID: 1 / Parent: 0 Details: Postsynaptic complexes were detected by Morse density tracing and classified by Affinity propagation (both implemented in PySeg) to obtain putative AMPA receptor particles. Afterwards the ...Details: Postsynaptic complexes were detected by Morse density tracing and classified by Affinity propagation (both implemented in PySeg) to obtain putative AMPA receptor particles. Afterwards the averaged were further refined by 3D classification in Relion. |
|---|---|
| Source (natural) | Organism:   Rattus norvegicus (Norway rat) / Organ: brain / Tissue: neocortex / Organelle: excitatory synapse / Location in cell: postsynaptic Rattus norvegicus (Norway rat) / Organ: brain / Tissue: neocortex / Organelle: excitatory synapse / Location in cell: postsynaptic |
| Molecular weight | Theoretical: 400 KDa |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Component - Name: HEPES-buffered saline Details: 140 mM NaCl, 5 mM KCl, 5 mM NaHCO3, 1.2 mM Na2HPO4, 1 mM MgCl2, 10 mM glucose, and 10 mM Hepes |
| Grid | Model: Quantifoil / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 300 K / Instrument: HOMEMADE PLUNGER |
| Details | Neocortical synaptosomal fraction |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.0 µm / Nominal defocus min: 0.5 µm Bright-field microscopy / Nominal defocus max: 1.0 µm / Nominal defocus min: 0.5 µm |
| Specialist optics | Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 1.66 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller