+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11212 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of ESCRT-III helical Vps24 filaments | |||||||||
 Map data Map data | Sharpened cryo EM density | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationEndosomal Sorting Complex Required For Transport (ESCRT) / intralumenal vesicle formation / Sealing of the nuclear envelope (NE) by ESCRT-III /  Macroautophagy / ATP export / Macroautophagy / ATP export /  ESCRT III complex / endosome transport via multivesicular body sorting pathway / late endosome to vacuole transport / ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway / ESCRT III complex / endosome transport via multivesicular body sorting pathway / late endosome to vacuole transport / ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway /  multivesicular body ...Endosomal Sorting Complex Required For Transport (ESCRT) / intralumenal vesicle formation / Sealing of the nuclear envelope (NE) by ESCRT-III / multivesicular body ...Endosomal Sorting Complex Required For Transport (ESCRT) / intralumenal vesicle formation / Sealing of the nuclear envelope (NE) by ESCRT-III /  Macroautophagy / ATP export / Macroautophagy / ATP export /  ESCRT III complex / endosome transport via multivesicular body sorting pathway / late endosome to vacuole transport / ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway / ESCRT III complex / endosome transport via multivesicular body sorting pathway / late endosome to vacuole transport / ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway /  multivesicular body / multivesicular body /  protein transport / identical protein binding / protein transport / identical protein binding /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Saccharomyces cerevisiae (brewer's yeast) / Saccharomyces cerevisiae (brewer's yeast) /   Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) | |||||||||
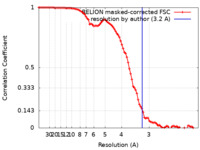
| Method | helical reconstruction /  cryo EM / Resolution: 3.2 Å cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Huber ST / Mostafavi S / Mortensen SA / Sachse C | |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2020 Journal: Sci Adv / Year: 2020Title: Structure and assembly of ESCRT-III helical Vps24 filaments. Authors: Stefan T Huber / Siavash Mostafavi / Simon A Mortensen / Carsten Sachse /  Abstract: ESCRT-III proteins mediate a range of cellular membrane remodeling activities such as multivesicular body biogenesis, cytokinesis, and viral release. Critical to these processes is the assembly of ...ESCRT-III proteins mediate a range of cellular membrane remodeling activities such as multivesicular body biogenesis, cytokinesis, and viral release. Critical to these processes is the assembly of ESCRT-III subunits into polymeric structures. In this study, we determined the cryo-EM structure of a helical assembly of Vps24 at 3.2-Å resolution and found that Vps24 adopts an elongated open conformation. Vps24 forms a domain-swapped dimer extended into protofilaments that associate into a double-stranded apolar filament. We demonstrate that, upon binding negatively charged lipids, Vps24 homopolymer filaments undergo partial disassembly into shorter filament fragments and oligomers. Upon the addition of Vps24, Vps2, and Snf7, liposomes are deformed into neck and tubular structures by an ESCRT-III heteropolymer coat. The filamentous Vps24 homopolymer assembly structure and interaction studies reveal how Vps24 could introduce unique geometric properties to mixed-type ESCRT-III heteropolymers and contribute to the process of membrane scission events. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11212.map.gz emd_11212.map.gz | 19.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11212-v30.xml emd-11212-v30.xml emd-11212.xml emd-11212.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11212_fsc.xml emd_11212_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_11212.png emd_11212.png | 204.5 KB | ||
| Masks |  emd_11212_msk_1.map emd_11212_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_11212_additional.map.gz emd_11212_additional.map.gz emd_11212_half_map_1.map.gz emd_11212_half_map_1.map.gz emd_11212_half_map_2.map.gz emd_11212_half_map_2.map.gz | 19.2 MB 49.6 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11212 http://ftp.pdbj.org/pub/emdb/structures/EMD-11212 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11212 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11212 | HTTPS FTP |
-Related structure data
| Related structure data |  6zh3MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10495 (Title: Structure and assembly of ESCRT-III helical Vps24 filaments EMPIAR-10495 (Title: Structure and assembly of ESCRT-III helical Vps24 filamentsData size: 70.0 Data #1: Aligned micrographs of Vps24 helical filaments [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11212.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11212.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened cryo EM density | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11212_msk_1.map emd_11212_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened cryo EM density
| File | emd_11212_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened cryo EM density | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_11212_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_11212_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Doubled-stranded helical filament assembly of Vps24
| Entire | Name: Doubled-stranded helical filament assembly of Vps24 |
|---|---|
| Components |
|
-Supramolecule #1: Doubled-stranded helical filament assembly of Vps24
| Supramolecule | Name: Doubled-stranded helical filament assembly of Vps24 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Macromolecule #1: Vacuolar protein-sorting-associated protein 24
| Macromolecule | Name: Vacuolar protein-sorting-associated protein 24 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast)Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 26.562174 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: GSHMDYIKKA IWGPDPKEQQ RRIRSVLRKN GRNIEKSLRE LTVLQNKTQQ LIKKSAKKND VRTVRLYAKE LYQINKQYDR MYTSRAQLD SVRMKIDEAI RMNTLSNQMA DSAGLMREVN SLVRLPQLRN TMIELEKELM KSGIISEMVD DTMESVGDVG E EMDEAVDE ...String: GSHMDYIKKA IWGPDPKEQQ RRIRSVLRKN GRNIEKSLRE LTVLQNKTQQ LIKKSAKKND VRTVRLYAKE LYQINKQYDR MYTSRAQLD SVRMKIDEAI RMNTLSNQMA DSAGLMREVN SLVRLPQLRN TMIELEKELM KSGIISEMVD DTMESVGDVG E EMDEAVDE EVNKIVEQYT NEKFKNVDQV PTVELAANEE EQEIPDEKVD EEADRMVNEM RERLRALQN |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.9 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Support film - Film thickness: 12.0 nm / Pretreatment - Type: GLOW DISCHARGE | |||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV | |||||||||
| Details | Sample was concentrated to 5 mg/mL, incubated in the refridgerator overnight, ultracentrifugated, and resuspended to a final concentration of 0.9 mg/mL for cryo-EM |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.75 µm / Nominal magnification: 130000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.75 µm / Nominal magnification: 130000 |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Number real images: 3257 / Average electron dose: 1.0 e/Å2 Details: 1320 images were manually selected for further processing |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z
Z Y
Y X
X