+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11172 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Disulfide-locked early prepore intermedilysin-CD59 | |||||||||
 Map data Map data | Half map 1 of ILY-CD59 early prepore oligomer formed on a lipid nanodsic, 5 subunits | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | early prepore /  membrane-bound / membrane-bound /  oligomer / oligomer /  TOXIN TOXIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of activation of membrane attack complex / complement binding /  regulation of complement-dependent cytotoxicity / regulation of complement-dependent cytotoxicity /  regulation of complement activation / Cargo concentration in the ER / COPII-mediated vesicle transport / regulation of complement activation / Cargo concentration in the ER / COPII-mediated vesicle transport /  cholesterol binding / tertiary granule membrane / specific granule membrane / COPI-mediated anterograde transport ...negative regulation of activation of membrane attack complex / complement binding / cholesterol binding / tertiary granule membrane / specific granule membrane / COPI-mediated anterograde transport ...negative regulation of activation of membrane attack complex / complement binding /  regulation of complement-dependent cytotoxicity / regulation of complement-dependent cytotoxicity /  regulation of complement activation / Cargo concentration in the ER / COPII-mediated vesicle transport / regulation of complement activation / Cargo concentration in the ER / COPII-mediated vesicle transport /  cholesterol binding / tertiary granule membrane / specific granule membrane / COPI-mediated anterograde transport / cholesterol binding / tertiary granule membrane / specific granule membrane / COPI-mediated anterograde transport /  transport vesicle / endoplasmic reticulum-Golgi intermediate compartment membrane / transport vesicle / endoplasmic reticulum-Golgi intermediate compartment membrane /  Regulation of Complement cascade / ER to Golgi transport vesicle membrane / Regulation of Complement cascade / ER to Golgi transport vesicle membrane /  blood coagulation / blood coagulation /  toxin activity / killing of cells of another organism / vesicle / cell surface receptor signaling pathway / external side of plasma membrane / toxin activity / killing of cells of another organism / vesicle / cell surface receptor signaling pathway / external side of plasma membrane /  Golgi membrane / Golgi membrane /  focal adhesion / Neutrophil degranulation / endoplasmic reticulum membrane / host cell plasma membrane / focal adhesion / Neutrophil degranulation / endoplasmic reticulum membrane / host cell plasma membrane /  cell surface / cell surface /  extracellular space / extracellular exosome / extracellular region / extracellular space / extracellular exosome / extracellular region /  membrane / membrane /  metal ion binding / metal ion binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Streptococcus intermedius (bacteria) / Streptococcus intermedius (bacteria) /   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.6 Å cryo EM / Resolution: 4.6 Å | |||||||||
 Authors Authors | Shah NR / Bubeck D | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Structural basis for tuning activity and membrane specificity of bacterial cytolysins. Authors: Nita R Shah / Tomas B Voisin / Edward S Parsons / Courtney M Boyd / Bart W Hoogenboom / Doryen Bubeck /  Abstract: Cholesterol-dependent cytolysins (CDCs) are pore-forming proteins that serve as major virulence factors for pathogenic bacteria. They target eukaryotic cells using different mechanisms, but all ...Cholesterol-dependent cytolysins (CDCs) are pore-forming proteins that serve as major virulence factors for pathogenic bacteria. They target eukaryotic cells using different mechanisms, but all require the presence of cholesterol to pierce lipid bilayers. How CDCs use cholesterol to selectively lyse cells is essential for understanding virulence strategies of several pathogenic bacteria, and for repurposing CDCs to kill new cellular targets. Here we address that question by trapping an early state of pore formation for the CDC intermedilysin, bound to the human immune receptor CD59 in a nanodisc model membrane. Our cryo electron microscopy map reveals structural transitions required for oligomerization, which include the lateral movement of a key amphipathic helix. We demonstrate that the charge of this helix is crucial for tuning lytic activity of CDCs. Furthermore, we discover modifications that overcome the requirement of cholesterol for membrane rupture, which may facilitate engineering the target-cell specificity of pore-forming proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11172.map.gz emd_11172.map.gz | 8.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11172-v30.xml emd-11172-v30.xml emd-11172.xml emd-11172.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_11172.png emd_11172.png | 146 KB | ||
| Filedesc metadata |  emd-11172.cif.gz emd-11172.cif.gz | 6.5 KB | ||
| Others |  emd_11172_half_map_1.map.gz emd_11172_half_map_1.map.gz emd_11172_half_map_2.map.gz emd_11172_half_map_2.map.gz | 9.7 MB 9.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11172 http://ftp.pdbj.org/pub/emdb/structures/EMD-11172 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11172 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11172 | HTTPS FTP |
-Related structure data
| Related structure data |  6zd0MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11172.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11172.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 of ILY-CD59 early prepore oligomer formed on a lipid nanodsic, 5 subunits | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half map 2 of ILY-CD59 early prepore oligomer...
| File | emd_11172_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 of ILY-CD59 early prepore oligomer formed on a lipid nanodsic, 5 subunits | ||||||||||||
| Projections & Slices |
| ||||||||||||
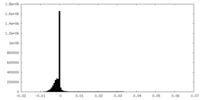
| Density Histograms |
-Half map: Half map 2 of ILY-CD59 early prepore oligomer...
| File | emd_11172_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 of ILY-CD59 early prepore oligomer formed on a lipid nanodsic, 5 subunits | ||||||||||||
| Projections & Slices |
| ||||||||||||
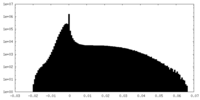
| Density Histograms |
- Sample components
Sample components
-Entire : Early prepore of intermedilysin-CD59
| Entire | Name: Early prepore of intermedilysin-CD59 |
|---|---|
| Components |
|
-Supramolecule #1: Early prepore of intermedilysin-CD59
| Supramolecule | Name: Early prepore of intermedilysin-CD59 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Prepore is formed on a lipid bilayer on a nanodisc |
|---|
-Supramolecule #2: Thiol-activated cytolysin
| Supramolecule | Name: Thiol-activated cytolysin / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Streptococcus intermedius (bacteria) Streptococcus intermedius (bacteria) |
-Supramolecule #3: CD59 glycoprotein
| Supramolecule | Name: CD59 glycoprotein / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Thiol-activated cytolysin
| Macromolecule | Name: Thiol-activated cytolysin / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Streptococcus intermedius (bacteria) Streptococcus intermedius (bacteria) |
| Molecular weight | Theoretical: 59.100953 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: MGGSHHHHHH GMASMTGGQQ MGRDLYDDDD KDRWGSETPT KPKAAQTEKK TEKKPENSNS EAAKKALNDY IWGLQYDKLN ILTHQGEKL KNHSSREAFH RPGEYVVCEK KKQSISNATS KLSVSSANDD RIFPGALLKA DQSLLENLPT LIPVNRGKTT I SVNLPGLK ...String: MGGSHHHHHH GMASMTGGQQ MGRDLYDDDD KDRWGSETPT KPKAAQTEKK TEKKPENSNS EAAKKALNDY IWGLQYDKLN ILTHQGEKL KNHSSREAFH RPGEYVVCEK KKQSISNATS KLSVSSANDD RIFPGALLKA DQSLLENLPT LIPVNRGKTT I SVNLPGLK NGESNLTVEN PSNSTVRTAV NNLVEKWIQN YSKTHAVPAR MQYESISAQS MSQLQAKFGA DFSKVGAPLN VD FSSVHKC EKQVFIANFR QVYYTASVDS PNSPSALFGS GITPTDLINR GVNSKTPPVY VSNVSYGRAM YVKFETTSKS TKV QAAIDA VVKGAKLKAG TEYENILKNT KITAVVLGGN PGEASKVITG NIDTLKDLIQ KGSNFSAQSP AVPISYTTSF VKDN SIATI QNNTDYIETK VTSYKDGALT LNHDGAFVAR FYVYWEELGH DADGYETIRS RSWSGNGYNR GAHYSTTLRF KGNVR NIRV KVLGATGLAW EPWRLIYSKN DLPLVPQRNI STWGTTLHPQ FEDKVVKDNT D UniProtKB:  Thiol-activated cytolysin Thiol-activated cytolysin |
-Macromolecule #2: CD59 glycoprotein
| Macromolecule | Name: CD59 glycoprotein / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 9.204445 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MLQCYNCPNP TADCKTAVNC SSDFDACLIT KAGLQVYNKC WKFEHCNFND VTTRLRENEL TYYCCKKDLC NFNEQLENC UniProtKB: CD59 glycoprotein |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR Details: Grids were glow discharged before application of graphene oxide, then left to dry for 1 hour before use. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 294 K / Instrument: FEI VITROBOT MARK III / Details: Wait time of 60 s, blot time 2.5 s, blot force 3. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.1 µm / Nominal defocus min: 1.9000000000000001 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.1 µm / Nominal defocus min: 1.9000000000000001 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Details | Pixel size 1.4 A |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average exposure time: 1.0 sec. / Average electron dose: 66.0 e/Å2 Details: Some images were collected at a 30 degree tilt. Images were collected in movie mode at 39 frames per second. |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL In silico model: A previous data set was used to generate a 8.6 A reconstruction, which was used as the startup model. |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION |
| Final 3D classification | Number classes: 4 / Software - Name: RELION |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION |
| Final reconstruction | Number classes used: 2 / Resolution.type: BY AUTHOR / Resolution: 4.6 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION / Number images used: 51041 |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Model of ILY-CD59 was fit and refined into the central subunit density. A combination of rigid body fitting and global minimization with secondary structure restraints was used to fit and refine the central subunit model. Then the central subunit model was rigid body fit as one body into the neighbouring subunits to generate a 3 subunit oligomer model. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-6zd0: |
 Movie
Movie Controller
Controller


















 Z
Z Y
Y X
X