[English] 日本語
 Yorodumi
Yorodumi- EMDB-11123: Pre-fusion conformation of glycoprotein B of Herpes simplex virus 1 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11123 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Pre-fusion conformation of glycoprotein B of Herpes simplex virus 1 | |||||||||||||||
 Map data Map data | Pre-fusion structure of glycoprotein B of Herpes simplex virus 1 | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell Golgi membrane / host cell endosome membrane / symbiont entry into host cell /  viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane /  membrane / identical protein binding membrane / identical protein bindingSimilarity search - Function | |||||||||||||||
| Biological species |    Human herpesvirus 1 (Herpes simplex virus type 1) / Human herpesvirus 1 (Herpes simplex virus type 1) /    Human alphaherpesvirus 1 (Herpes simplex virus type 1) Human alphaherpesvirus 1 (Herpes simplex virus type 1) | |||||||||||||||
| Method | subtomogram averaging /  cryo EM / Resolution: 9.1 Å cryo EM / Resolution: 9.1 Å | |||||||||||||||
 Authors Authors | Vollmer B / Prazak V / Vasishtan D / Jefferys EE / Hernandez-Duran A / Vallbracht M / Klupp B / Mettenleiter TC / Backovic M / Rey FA ...Vollmer B / Prazak V / Vasishtan D / Jefferys EE / Hernandez-Duran A / Vallbracht M / Klupp B / Mettenleiter TC / Backovic M / Rey FA / Topf M / Gruenewald K | |||||||||||||||
| Funding support |  Germany, Germany,  United Kingdom, European Union, 4 items United Kingdom, European Union, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2020 Journal: Sci Adv / Year: 2020Title: The prefusion structure of herpes simplex virus glycoprotein B. Authors: B Vollmer / V Pražák / D Vasishtan / E E Jefferys / A Hernandez-Duran / M Vallbracht / B G Klupp / T C Mettenleiter / M Backovic / F A Rey / M Topf / K Grünewald /    Abstract: Cell entry of enveloped viruses requires specialized viral proteins that mediate fusion with the host membrane by substantial structural rearrangements from a metastable pre- to a stable postfusion ...Cell entry of enveloped viruses requires specialized viral proteins that mediate fusion with the host membrane by substantial structural rearrangements from a metastable pre- to a stable postfusion conformation. This metastability renders the herpes simplex virus 1 (HSV-1) fusion glycoprotein B (gB) highly unstable such that it readily converts into the postfusion form, thereby precluding structural elucidation of the pharmacologically relevant prefusion conformation. By identification of conserved sequence signatures and molecular dynamics simulations, we devised a mutation that stabilized this form. Functionally locking gB allowed the structural determination of its membrane-embedded prefusion conformation at sub-nanometer resolution and enabled the unambiguous fit of all ectodomains. The resulting pseudo-atomic model reveals a notable conservation of conformational domain rearrangements during fusion between HSV-1 gB and the vesicular stomatitis virus glycoprotein G, despite their very distant phylogeny. In combination with our comparative sequence-structure analysis, these findings suggest common fusogenic domain rearrangements in all class III viral fusion proteins. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11123.map.gz emd_11123.map.gz | 3.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11123-v30.xml emd-11123-v30.xml emd-11123.xml emd-11123.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
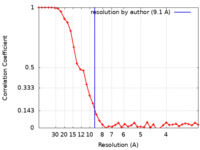
| FSC (resolution estimation) |  emd_11123_fsc.xml emd_11123_fsc.xml | 4.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_11123.png emd_11123.png | 158.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11123 http://ftp.pdbj.org/pub/emdb/structures/EMD-11123 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11123 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11123 | HTTPS FTP |
-Related structure data
| Related structure data |  6z9mMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11123.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11123.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Pre-fusion structure of glycoprotein B of Herpes simplex virus 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.6 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human alphaherpesvirus 1
| Entire | Name:    Human alphaherpesvirus 1 (Herpes simplex virus type 1) Human alphaherpesvirus 1 (Herpes simplex virus type 1) |
|---|---|
| Components |
|
-Supramolecule #1: Human alphaherpesvirus 1
| Supramolecule | Name: Human alphaherpesvirus 1 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Protein recombinantly expressed in membrane protein enriched extracellular vesicles (MPEEVs) NCBI-ID: 10298 / Sci species name: Human alphaherpesvirus 1 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: OTHER / Virus enveloped: Yes / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Host system | Organism:   Mesocricetus auratus (golden hamster) / Recombinant cell: Fibroblast Mesocricetus auratus (golden hamster) / Recombinant cell: Fibroblast |
| Molecular weight | Theoretical: 300 KDa |
-Macromolecule #1: Envelope glycoprotein B
| Macromolecule | Name: Envelope glycoprotein B / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Human herpesvirus 1 (Herpes simplex virus type 1) Human herpesvirus 1 (Herpes simplex virus type 1) |
| Molecular weight | Theoretical: 100.383305 KDa |
| Recombinant expression | Organism:   Mesocricetus auratus (golden hamster) Mesocricetus auratus (golden hamster) |
| Sequence | String: MHQGAPSWGR RWFVVWALLG LTLGVLVASA APSSPGTPGV AAATQAANGG PATPAPPALG AAPTGDPKPK KNKKPKNPTP PRPAGDNAT VAAGHATLRE HLRDIKAENT DANFYVCPPP TGATVVQFEQ PRRCPTRPEG QNYTEGIAVV FKENIAPYKF K ATMYYKDV ...String: MHQGAPSWGR RWFVVWALLG LTLGVLVASA APSSPGTPGV AAATQAANGG PATPAPPALG AAPTGDPKPK KNKKPKNPTP PRPAGDNAT VAAGHATLRE HLRDIKAENT DANFYVCPPP TGATVVQFEQ PRRCPTRPEG QNYTEGIAVV FKENIAPYKF K ATMYYKDV TVSQVWFGHR YSQFMGIFED RAPVPFEEVI DKINAKGVCR STAKYVRNNL ETTAFHRDDH ETDMELKPAN AA TRTSRGW HTTDLKYNPS RVEAFHRYGT TVNCIVEEVD ARSVYPYDEF VLATGDFVYM SPFYGYREGS HTEHTSYAAD RFK QVDGFY ARDLTTKARA TAPTTRNLLT TPKFTVAWDW VPKRPSVCTM TKWQEVDEML RSEYGGSFRF SSDAISTTFT TNLT EYPLS RVDLGDCIGK DARDAMDRIF ARRYNATHIK VGQPQYYLAN GGFLIAYQPL LSNTLAELYV REHLREQSRK PPNPT PPPP GASANASVER IKTTSSIEFA RLQFTYNHIQ RPVNDMLGRV AIAWCELQNH ELTLWNEARK LNPNAIASVT VGRRVS ARM LGDVMAVSTC VPVAADNVIV QNSMRISSRP GACYSRPLVS FRYEDQGPLV EGQLGENNEL RLTRDAIEPC TVGHRRY FT FGGGYVYFEE YAYSHQLSRA DITTVSTFID LNITMLEDHE FVPLEVYTRH EIKDSGLLDY TEVQRRNQLH DLRFADID T VIHADANAAM FAGLGAFFEG MGDLGRAVGK VVMGIVGGVV SAVSGVSSFM SNPFGALAVG LLVLAGLAAA FFAFRYVMR LQSNPMKALY PLTTKELKNP TNPDASGEGE EGGDFDEAKL AEAREMIRYM ALVSAMERTE HKAKKKGTSA LLSAKVTDMV MRKRRNTNY TQVPNKDGDA DEDDL |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 12.0 nm / Pretreatment - Type: GLOW DISCHARGE | |||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Instrument: HOMEMADE PLUNGER | |||||||||
| Details | Protein recombinantly expressed in membrane protein enriched extracellular vesicles (MPEEVs) |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 3.5 µm / Calibrated defocus min: 1.6 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.0 mm Bright-field microscopy / Cs: 2.0 mm |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
| Details | Additional Dataset collected using Titan Krios (FEI Thermo) at 300 kV with a 70 um C2 aperture and post-column QUANTUM energy filter operated in Zero-Loss mode using 20 eV energy slit and K2 Summit direct electron detector in counting mode (Gatan). Defocus range: 2300 - 4900 nm. |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3836 pixel / Digitization - Dimensions - Height: 3710 pixel / Average electron dose: 2.3 e/Å2 |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Residue range: 104-723 |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 300 / Target criteria: Cross correlation coefficient |
| Output model |  PDB-6z9m: |
 Movie
Movie Controller
Controller