[English] 日本語
 Yorodumi
Yorodumi- EMDB-10893: Structure of GldLM, the proton-powered motor that drives protein ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10893 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of GldLM, the proton-powered motor that drives protein transport and gliding motility | ||||||||||||
 Map data Map data | post processed volume | ||||||||||||
 Sample Sample |
| ||||||||||||
| Function / homology |  Function and homology information Function and homology information | ||||||||||||
| Biological species |  Flavobacterium johnsoniae (bacteria) Flavobacterium johnsoniae (bacteria) | ||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.9 Å cryo EM / Resolution: 3.9 Å | ||||||||||||
 Authors Authors | Hennell James R / Deme JC / Lea SM | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2020 Journal: Nat Microbiol / Year: 2020Title: Structure of a proton-powered molecular motor that drives protein transport and gliding motility Authors: Hennell James R / Deme JC / Kjaer A / Alcock F / Silale A / Lauber F / Bersk BC / Lea SM | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10893.map.gz emd_10893.map.gz | 5.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10893-v30.xml emd-10893-v30.xml emd-10893.xml emd-10893.xml | 22.3 KB 22.3 KB | Display Display |  EMDB header EMDB header |
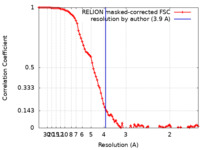
| FSC (resolution estimation) |  emd_10893_fsc.xml emd_10893_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_10893.png emd_10893.png | 167.6 KB | ||
| Masks |  emd_10893_msk_1.map emd_10893_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_10893_additional_1.map.gz emd_10893_additional_1.map.gz emd_10893_additional_2.map.gz emd_10893_additional_2.map.gz emd_10893_half_map_1.map.gz emd_10893_half_map_1.map.gz emd_10893_half_map_2.map.gz emd_10893_half_map_2.map.gz | 49.4 MB 42.2 MB 49.6 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10893 http://ftp.pdbj.org/pub/emdb/structures/EMD-10893 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10893 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10893 | HTTPS FTP |
-Related structure data
| Related structure data |  6ys8MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10893.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10893.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | post processed volume | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.822 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10893_msk_1.map emd_10893_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
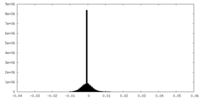
| Density Histograms |
-Additional map: 3D refinement volume
| File | emd_10893_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D refinement volume | ||||||||||||
| Projections & Slices |
| ||||||||||||
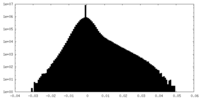
| Density Histograms |
-Additional map: RELION local resolution filtered volume
| File | emd_10893_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION local resolution filtered volume | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map
| File | emd_10893_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
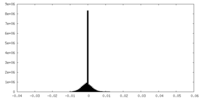
| Density Histograms |
-Half map: half map
| File | emd_10893_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
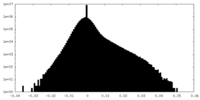
| Density Histograms |
- Sample components
Sample components
-Entire : GldLM
| Entire | Name: GldLM |
|---|---|
| Components |
|
-Supramolecule #1: GldLM
| Supramolecule | Name: GldLM / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Flavobacterium johnsoniae (bacteria) Flavobacterium johnsoniae (bacteria) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Macromolecule #1: GldM
| Macromolecule | Name: GldM / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Flavobacterium johnsoniae (bacteria) Flavobacterium johnsoniae (bacteria) |
| Molecular weight | Theoretical: 54.8565 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MAGGKLTPRQ KMINLMYLVF IAMLAMNVSK EVISAFGLMN EKFEAANTSS VTTNESLLTS LDQKAAEAKG EFAKAAETAH KVQAASKEF YDYIGTLKTQ AVKGFEVDKE TGKMPYEAMD RGDNIDDWFT GDGYTKKGNE IIAKIEKYKS DIKAALGTDK K YAGIISEV ...String: MAGGKLTPRQ KMINLMYLVF IAMLAMNVSK EVISAFGLMN EKFEAANTSS VTTNESLLTS LDQKAAEAKG EFAKAAETAH KVQAASKEF YDYIGTLKTQ AVKGFEVDKE TGKMPYEAMD RGDNIDDWFT GDGYTKKGNE IIAKIEKYKS DIKAALGTDK K YAGIISEV EKKFDVSDVK NKEGIKEKYL AYHFKGFPAI ASAAKLSAWQ NDVKKTEADV YNSALGKAAV AAASYSNYQA IV VLDKNAY FQGEKVTGKV VLGRYDENTK PTSFQGPGQI VNGQAVISLT AGGVGEQDIN GQFTFLEDGK NIPLKFSGKY VVV PRPNSA TISADKMNVV YRGVVNPISV SFAGVDANKI VASAPGLSSA GKPGKYNMSP GQGTEATISV TGTLPNGDKV TDKK TFRIK GIPGPTGTIR GEMGVVKGPK SNLEIATIGA KLLDFDFEVG LDVVGFNMKI AGQPTVVVTG NKMNAQCKSV LARAG KGDQ VTISEIKTKL VGAGSYLLPR TAPVIYEIQ |
-Macromolecule #2: GldL
| Macromolecule | Name: GldL / type: protein_or_peptide / ID: 2 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Flavobacterium johnsoniae (bacteria) Flavobacterium johnsoniae (bacteria) |
| Molecular weight | Theoretical: 22.786209 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MALLSKKVMN FAYGMGAAVV IVGALFKITH FEIGPLTGTV MLSIGLLTEA LIFALSAFEP VEDELDWTLV YPELANGQAR KKEAKAETA TDAQGLLSQK LDAMLKEAKV DGELMASLGN SIKNFEGAAK AISPTVDSIA GQKKYAEEMS MAAAQMESLN S LYKVQLES ...String: MALLSKKVMN FAYGMGAAVV IVGALFKITH FEIGPLTGTV MLSIGLLTEA LIFALSAFEP VEDELDWTLV YPELANGQAR KKEAKAETA TDAQGLLSQK LDAMLKEAKV DGELMASLGN SIKNFEGAAK AISPTVDSIA GQKKYAEEMS MAAAQMESLN S LYKVQLES ASRNAQANSE IAENAAKLKE QMASMTANIA SLNSVYGGML SAMSNKG |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER | ||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 0.0003 µm / Nominal defocus min: 0.0001 µm / Nominal magnification: 165000 Bright-field microscopy / Nominal defocus max: 0.0003 µm / Nominal defocus min: 0.0001 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 48.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6ys8: |
 Movie
Movie Controller
Controller













 Z
Z Y
Y X
X