[English] 日本語
 Yorodumi
Yorodumi- EMDB-10813: In-situ structure of a dimer of the trimeric HEF from influenza C... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10813 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | In-situ structure of a dimer of the trimeric HEF from influenza C viral particles without a matrix layer by cryo-ET | ||||||||||||
 Map data Map data | Post-processed map. Dimer of trimers from virions without a matrix layer | ||||||||||||
 Sample Sample |
| ||||||||||||
| Function / homology |  Function and homology information Function and homology informationsialate 9-O-acetylesterase activity / sialate 4-O-acetylesterase activity /  sialate O-acetylesterase / sialate O-acetylesterase /  viral budding from plasma membrane / endocytosis involved in viral entry into host cell / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral budding from plasma membrane / endocytosis involved in viral entry into host cell / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope / virion attachment to host cell ...sialate 9-O-acetylesterase activity / sialate 4-O-acetylesterase activity / viral envelope / virion attachment to host cell ...sialate 9-O-acetylesterase activity / sialate 4-O-acetylesterase activity /  sialate O-acetylesterase / sialate O-acetylesterase /  viral budding from plasma membrane / endocytosis involved in viral entry into host cell / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral budding from plasma membrane / endocytosis involved in viral entry into host cell / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane /  membrane membraneSimilarity search - Function | ||||||||||||
| Biological species |   Canis lupus (gray wolf) / Canis lupus (gray wolf) /   Influenza C virus Influenza C virus | ||||||||||||
| Method | subtomogram averaging /  cryo EM / Resolution: 11.3 Å cryo EM / Resolution: 11.3 Å | ||||||||||||
 Authors Authors | Halldorsson S / Rosenthal PB | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: In situ structure and organization of the influenza C virus surface glycoprotein. Authors: Steinar Halldorsson / Kasim Sader / Jack Turner / Lesley J Calder / Peter B Rosenthal /   Abstract: The lipid-enveloped influenza C virus contains a single surface glycoprotein, the haemagglutinin-esterase-fusion (HEF) protein, that mediates receptor binding, receptor destruction, and membrane ...The lipid-enveloped influenza C virus contains a single surface glycoprotein, the haemagglutinin-esterase-fusion (HEF) protein, that mediates receptor binding, receptor destruction, and membrane fusion at the low pH of the endosome. Here we apply electron cryotomography and subtomogram averaging to describe the structural basis for hexagonal lattice formation by HEF on the viral surface. The conformation of the glycoprotein in situ is distinct from the structure of the isolated trimeric ectodomain, showing that a splaying of the membrane distal domains is required to mediate contacts that form the lattice. The splaying of these domains is also coupled to changes in the structure of the stem region which is involved in membrane fusion, thereby linking HEF's membrane fusion conformation with its assembly on the virus surface. The glycoprotein lattice can form independent of other virion components but we show a major role for the matrix layer in particle formation. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10813.map.gz emd_10813.map.gz | 28.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10813-v30.xml emd-10813-v30.xml emd-10813.xml emd-10813.xml | 20.7 KB 20.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10813_fsc.xml emd_10813_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_10813.png emd_10813.png | 133.3 KB | ||
| Masks |  emd_10813_msk_1.map emd_10813_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Others |  emd_10813_additional_1.map.gz emd_10813_additional_1.map.gz emd_10813_half_map_1.map.gz emd_10813_half_map_1.map.gz emd_10813_half_map_2.map.gz emd_10813_half_map_2.map.gz | 22.4 MB 22.5 MB 22.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10813 http://ftp.pdbj.org/pub/emdb/structures/EMD-10813 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10813 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10813 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10813.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10813.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post-processed map. Dimer of trimers from virions without a matrix layer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10813_msk_1.map emd_10813_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
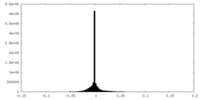
| Density Histograms |
-Additional map: Unfiltered map
| File | emd_10813_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered map | ||||||||||||
| Projections & Slices |
| ||||||||||||
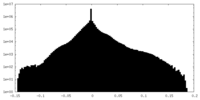
| Density Histograms |
-Half map: Half map 1
| File | emd_10813_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
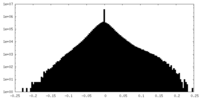
| Density Histograms |
-Half map: Half map 2
| File | emd_10813_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Influenza C virus
| Entire | Name:   Influenza C virus Influenza C virus |
|---|---|
| Components |
|
-Supramolecule #1: Influenza C virus
| Supramolecule | Name: Influenza C virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Purified from infected cells. / NCBI-ID: 11552 / Sci species name: Influenza C virus / Sci species strain: C/Johannesburg/1/1966 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Hemagglutinin esterase fusion
| Macromolecule | Name: Hemagglutinin esterase fusion / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number:  sialate O-acetylesterase sialate O-acetylesterase |
|---|---|
| Source (natural) | Organism:   Canis lupus (gray wolf) Canis lupus (gray wolf) |
| Sequence | String: MFFSLLLVLG LTEAEKIKIC LQKQVNSSFS LHNGFGGNLY ATEEKRMFEL VKPKAGASVL NQSTWIGFG DSRTDKSNSA FPRSADVSAK TADKFRFLSG GSLMLSMFGP PGKVDYLYQG C GKHKVFYE GVNWSPHAAI NCYRKNWTDI KLNFQKNIYE LASQSHCMSL ...String: MFFSLLLVLG LTEAEKIKIC LQKQVNSSFS LHNGFGGNLY ATEEKRMFEL VKPKAGASVL NQSTWIGFG DSRTDKSNSA FPRSADVSAK TADKFRFLSG GSLMLSMFGP PGKVDYLYQG C GKHKVFYE GVNWSPHAAI NCYRKNWTDI KLNFQKNIYE LASQSHCMSL VNALDKTIPL QV TAGTAGN CNNSFLKNPA LYTQEVKPSE NKCGKENLAF FTLPTQFGTY ECKLHLVASC YFI YDSKEV YNKRGCDNYF QVIYDSFGKV VGGLDNRVSP YTGNSGDTPT MQCDMLQLKP GRYS VRSSP RFLLMPERSY CFDMKEKGPV TAVQSIWGKG RESDYAVDQA CLSTPGCMLI QKQKP YIGE ADDHHGDQEM RELLSGLDYE ARCISQSGWV NETSPFTEKY LLPPKFGRCP LAAKEE SIP KIPDGLLIPT SGTDTTVTKP KSRIFGIDDL IIGVLFVAIV ETGIGGYLLG SRKESGG GV TKESAEKGFE KIGNDIQILK SSINIAIEKL NDRISHDEQA IRDLTLEIEN ARSEALLG E LGIIRALLVG NISIGLQESL WELASEITNR AGDLAVEVSP GCWIIDNNIC DQSCQNFIF KFNETAPVPT IPPLDTKIDL QSDPFYWGSS LGLAITATIS LAALVISGIA ICRTK |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: Quantifoil / Support film - Material: CARBON / Support film - topology: HOLEY | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III | ||||||||||||
| Details | Heterogeneous mix of viral particles, viral like particles and cellular vesicles |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 0.0045000000000000005 µm / Nominal defocus min: 0.0025 µm / Nominal magnification: 64000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 0.0045000000000000005 µm / Nominal defocus min: 0.0025 µm / Nominal magnification: 64000 |
| Specialist optics | Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Number grids imaged: 2 / Average exposure time: 1.57 sec. / Average electron dose: 1.57 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
 Movie
Movie Controller
Controller



















 Z
Z Y
Y X
X