[English] 日本語
 Yorodumi
Yorodumi- EMDB-10765: Cryo-electron tomogram and segmentation of the cortical ER in yea... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10765 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-electron tomogram and segmentation of the cortical ER in yeast used for testing membrane curvature estimation algorithms. | ||||||||||||
 Map data Map data | Raw tomogram. | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) | ||||||||||||
| Method |  electron tomography / electron tomography /  cryo EM cryo EM | ||||||||||||
 Authors Authors | Collado JF / Salfer M | ||||||||||||
| Funding support |  Germany, 3 items Germany, 3 items
| ||||||||||||
 Citation Citation |  Journal: PLoS Comput Biol / Year: 2020 Journal: PLoS Comput Biol / Year: 2020Title: Reliable estimation of membrane curvature for cryo-electron tomography. Authors: Maria Salfer / Javier F Collado / Wolfgang Baumeister / Rubén Fernández-Busnadiego / Antonio Martínez-Sánchez /  Abstract: Curvature is a fundamental morphological descriptor of cellular membranes. Cryo-electron tomography (cryo-ET) is particularly well-suited to visualize and analyze membrane morphology in a close-to- ...Curvature is a fundamental morphological descriptor of cellular membranes. Cryo-electron tomography (cryo-ET) is particularly well-suited to visualize and analyze membrane morphology in a close-to-native state and molecular resolution. However, current curvature estimation methods cannot be applied directly to membrane segmentations in cryo-ET, as these methods cannot cope with some of the artifacts introduced during image acquisition and membrane segmentation, such as quantization noise and open borders. Here, we developed and implemented a Python package for membrane curvature estimation from tomogram segmentations, which we named PyCurv. From a membrane segmentation, a signed surface (triangle mesh) is first extracted. The triangle mesh is then represented by a graph, which facilitates finding neighboring triangles and the calculation of geodesic distances necessary for local curvature estimation. PyCurv estimates curvature based on tensor voting. Beside curvatures, this algorithm also provides robust estimations of surface normals and principal directions. We tested PyCurv and three well-established methods on benchmark surfaces and biological data. This revealed the superior performance of PyCurv not only for cryo-ET, but also for data generated by other techniques such as light microscopy and magnetic resonance imaging. Altogether, PyCurv is a versatile open-source software to reliably estimate curvature of membranes and other surfaces in a wide variety of applications. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10765.map.gz emd_10765.map.gz | 91.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10765-v30.xml emd-10765-v30.xml emd-10765.xml emd-10765.xml | 12.4 KB 12.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10765.png emd_10765.png | 239.7 KB | ||
| Masks |  emd_10765_msk_1.map emd_10765_msk_1.map | 12.6 MB |  Mask map Mask map | |
| Others |  emd_10765_additional.map.gz emd_10765_additional.map.gz emd_10765_additional_1.map.gz emd_10765_additional_1.map.gz | 113.1 MB 113.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10765 http://ftp.pdbj.org/pub/emdb/structures/EMD-10765 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10765 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10765 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10765.map.gz / Format: CCP4 / Size: 202 MB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) Download / File: emd_10765.map.gz / Format: CCP4 / Size: 202 MB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Raw tomogram. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 13.68 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10765_msk_1.map emd_10765_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
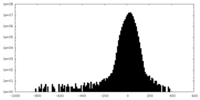
| Density Histograms |
-Additional map: Filtered tomogram using a deconvolution filter (https://github.com/dtegunov/tom deconv) executed...
| File | emd_10765_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Filtered tomogram using a deconvolution filter (https://github.com/dtegunov/tom_deconv) executed in MATLAB (Mathworks) using the functionalities of the TOM toolbox (Nickel et al. 2005). | ||||||||||||
| Projections & Slices |
| ||||||||||||
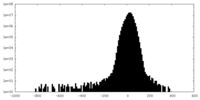
| Density Histograms |
-Additional map: Filtered tomogram using a deconvolution filter (https://github.com/dtegunov/tom deconv) executed...
| File | emd_10765_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Filtered tomogram using a deconvolution filter (https://github.com/dtegunov/tom_deconv) executed in MATLAB (Mathworks) using the functionalities of the TOM toolbox (Nickel et al. 2005). | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Saccharomyces cerevisiae strain ANDY129 delta Scs2 delta Scs22 de...
| Entire | Name: Saccharomyces cerevisiae strain ANDY129 delta Scs2 delta Scs22 delta Ist2 |
|---|---|
| Components |
|
-Supramolecule #1: Saccharomyces cerevisiae strain ANDY129 delta Scs2 delta Scs22 de...
| Supramolecule | Name: Saccharomyces cerevisiae strain ANDY129 delta Scs2 delta Scs22 delta Ist2 type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) |
| Recombinant expression | Organism:   Saccharomyces cerevisiae (brewer's yeast) / Recombinant strain: ANDY129 Saccharomyces cerevisiae (brewer's yeast) / Recombinant strain: ANDY129 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  electron tomography electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
| Sectioning | Focused ion beam - Instrument: OTHER / Focused ion beam - Ion: OTHER / Focused ion beam - Voltage: 30 kV / Focused ion beam - Current: 0.03 nA / Focused ion beam - Duration: 4000 sec. / Focused ion beam - Temperature: 93 K / Focused ion beam - Initial thickness: 1000 nm / Focused ion beam - Final thickness: 200 nm Focused ion beam - Details: The value given for _emd_sectioning_focused_ion_beam.instrument is Quanta 3D cryo-FIB / SEM. This is not in a list of allowed values {'DB235', 'OTHER'} so OTHER is written into the XML file. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.1 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 5.0 µm / Nominal magnification: 42000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 5.0 µm / Nominal magnification: 42000 |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Temperature | Min: 80.0 K / Max: 90.0 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average exposure time: 1.6 sec. / Average electron dose: 1.7 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Algorithm: BACK PROJECTION / Software - Name:  IMOD / Number images used: 52 IMOD / Number images used: 52 |
|---|
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X