[English] 日本語
 Yorodumi
Yorodumi- EMDB-10523: Cryo-EM structure of Toxoplasma gondii mitochondrial ATP synthase... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10523 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Toxoplasma gondii mitochondrial ATP synthase dimer, rotor-stator map | |||||||||||||||
 Map data Map data | Toxoplasma gondii ATP synthase dimer, rotor-stator full map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationmitochondrial proton-transporting ATP synthase complex, catalytic sector F(1) / proton-transporting ATP synthase complex, coupling factor F(o) / proton motive force-driven ATP synthesis / proton transmembrane transporter activity / proton-transporting ATP synthase complex, catalytic core F(1) / proton-transporting ATP synthase activity, rotational mechanism /  mitochondrial inner membrane / intracellular membrane-bounded organelle / mitochondrial inner membrane / intracellular membrane-bounded organelle /  lipid binding / lipid binding /  membrane / membrane /  cytoplasm cytoplasmSimilarity search - Function | |||||||||||||||
| Biological species |   Toxoplasma gondii GT1 (eukaryote) / Toxoplasma gondii GT1 (eukaryote) /   Toxoplasma gondii (strain ATCC 50853 / GT1) (eukaryote) Toxoplasma gondii (strain ATCC 50853 / GT1) (eukaryote) | |||||||||||||||
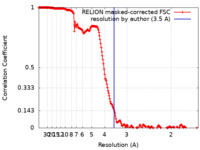
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.5 Å cryo EM / Resolution: 3.5 Å | |||||||||||||||
 Authors Authors | Muhleip A / Kock Flygaard R / Amunts A | |||||||||||||||
| Funding support |  Sweden, 4 items Sweden, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: ATP synthase hexamer assemblies shape cristae of Toxoplasma mitochondria. Authors: Alexander Mühleip / Rasmus Kock Flygaard / Jana Ovciarikova / Alice Lacombe / Paula Fernandes / Lilach Sheiner / Alexey Amunts /   Abstract: Mitochondrial ATP synthase plays a key role in inducing membrane curvature to establish cristae. In Apicomplexa causing diseases such as malaria and toxoplasmosis, an unusual cristae morphology has ...Mitochondrial ATP synthase plays a key role in inducing membrane curvature to establish cristae. In Apicomplexa causing diseases such as malaria and toxoplasmosis, an unusual cristae morphology has been observed, but its structural basis is unknown. Here, we report that the apicomplexan ATP synthase assembles into cyclic hexamers, essential to shape their distinct cristae. Cryo-EM was used to determine the structure of the hexamer, which is held together by interactions between parasite-specific subunits in the lumenal region. Overall, we identified 17 apicomplexan-specific subunits, and a minimal and nuclear-encoded subunit-a. The hexamer consists of three dimers with an extensive dimer interface that includes bound cardiolipins and the inhibitor IF. Cryo-ET and subtomogram averaging revealed that hexamers arrange into ~20-megadalton pentagonal pyramids in the curved apical membrane regions. Knockout of the linker protein ATPTG11 resulted in the loss of pentagonal pyramids with concomitant aberrantly shaped cristae. Together, this demonstrates that the unique macromolecular arrangement is critical for the maintenance of cristae morphology in Apicomplexa. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10523.map.gz emd_10523.map.gz | 378 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10523-v30.xml emd-10523-v30.xml emd-10523.xml emd-10523.xml | 21.3 KB 21.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10523_fsc.xml emd_10523_fsc.xml | 19.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_10523.png emd_10523.png | 39 KB | ||
| Masks |  emd_10523_msk_1.map emd_10523_msk_1.map | 669.9 MB |  Mask map Mask map | |
| Others |  emd_10523_half_map_1.map.gz emd_10523_half_map_1.map.gz emd_10523_half_map_2.map.gz emd_10523_half_map_2.map.gz | 538.6 MB 538.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10523 http://ftp.pdbj.org/pub/emdb/structures/EMD-10523 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10523 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10523 | HTTPS FTP |
-Related structure data
| Related structure data |  6tmjMC  6tmgC  6tmhC  6tmiC  6tmkC  6tmlC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10523.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10523.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Toxoplasma gondii ATP synthase dimer, rotor-stator full map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10523_msk_1.map emd_10523_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Halfmap 2
| File | emd_10523_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Halfmap 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Halfmap 1
| File | emd_10523_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Halfmap 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Mitochondrial ATP synthase dimer, rotor-stator
| Entire | Name: Mitochondrial ATP synthase dimer, rotor-stator |
|---|---|
| Components |
|
-Supramolecule #1: Mitochondrial ATP synthase dimer, rotor-stator
| Supramolecule | Name: Mitochondrial ATP synthase dimer, rotor-stator / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Toxoplasma gondii GT1 (eukaryote) Toxoplasma gondii GT1 (eukaryote) |
| Molecular weight | Theoretical: 127 KDa |
-Macromolecule #1: ATPTG11
| Macromolecule | Name: ATPTG11 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Toxoplasma gondii (strain ATCC 50853 / GT1) (eukaryote) Toxoplasma gondii (strain ATCC 50853 / GT1) (eukaryote)Strain: ATCC 50853 / GT1 |
| Molecular weight | Theoretical: 15.425367 KDa |
| Sequence | String: MVRNQRYPAS PVQEIFLPEP VPFVQFDQTA PSPNSPPAPL PSPSLSQCEE QKDRYRDISS MFHRGVAGAE QVREAYNSMA KCFRRVSVA EVLESDPAFR QARNFTMDLK QAEDDQRYKQ LQYGRVPSIL TKYHL |
-Macromolecule #2: subunit a
| Macromolecule | Name: subunit a / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Toxoplasma gondii (strain ATCC 50853 / GT1) (eukaryote) Toxoplasma gondii (strain ATCC 50853 / GT1) (eukaryote)Strain: ATCC 50853 / GT1 |
| Molecular weight | Theoretical: 24.56257 KDa |
| Sequence | String: MAAGSRFPFC TAARLSSRGT LPRLGEATFF AGAESQRSAG AFAKTLQRPF LRAPSTQLFP VGNRLGVSSA RALVANAMEP RRFFAAAAS AKATHALQPT GTGSVAFTRP GQGSNAQFQT SLADKTRGLL GVGFLRPTKM ASFAATFLLN FRFYFMYMAR T TFQAVRPL ...String: MAAGSRFPFC TAARLSSRGT LPRLGEATFF AGAESQRSAG AFAKTLQRPF LRAPSTQLFP VGNRLGVSSA RALVANAMEP RRFFAAAAS AKATHALQPT GTGSVAFTRP GQGSNAQFQT SLADKTRGLL GVGFLRPTKM ASFAATFLLN FRFYFMYMAR T TFQAVRPL LAFSVFGEVM KLVLATMSSG LFSFLFSFVL AFEVFYFFLQ CYISYTFLTM FFTVLF |
-Macromolecule #3: ATP synthase subunit gamma
| Macromolecule | Name: ATP synthase subunit gamma / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Toxoplasma gondii (strain ATCC 50853 / GT1) (eukaryote) Toxoplasma gondii (strain ATCC 50853 / GT1) (eukaryote)Strain: ATCC 50853 / GT1 |
| Molecular weight | Theoretical: 34.573031 KDa |
| Sequence | String: MAGLASLSSV GALRGMRLVP AAHLLPLHSA FGQQTRNFGA GDLKIVAARM KSVKSIQKIT KAMKMVAASK LRMDQRRLEN GLPFATPVQ KLVQRIPVDP KEKGTLAVLA LSSDKGLCGG VNSFVAKQAR IVIKENEMAG NAVQVYGVGD KIRSALQRTF G DRFKRIMT ...String: MAGLASLSSV GALRGMRLVP AAHLLPLHSA FGQQTRNFGA GDLKIVAARM KSVKSIQKIT KAMKMVAASK LRMDQRRLEN GLPFATPVQ KLVQRIPVDP KEKGTLAVLA LSSDKGLCGG VNSFVAKQAR IVIKENEMAG NAVQVYGVGD KIRSALQRTF G DRFKRIMT EVTRFPWNFG QACIIADRLM QDNPARLMVI YNHFKSAVAY DTLTLNVLTP TQAAQSAKEQ LNTFEFEPEK TD VWKDLQD FYYACTVFGC MLDNIASEQS ARMSAMDNAS TNAGEMISSL TLRYNRARQA KITTELVEII SGANALE |
-Macromolecule #4: ATP synthase subunit delta
| Macromolecule | Name: ATP synthase subunit delta / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Toxoplasma gondii (strain ATCC 50853 / GT1) (eukaryote) Toxoplasma gondii (strain ATCC 50853 / GT1) (eukaryote)Strain: ATCC 50853 / GT1 |
| Molecular weight | Theoretical: 19.476082 KDa |
| Sequence | String: MFARAFSRFA SLAAPAPQRG WNAFVLPSRH FATAAGGANP FKNQLLLTLS SPSEAIYVRT PVRSVTVPGS EGAMTMTNGH SQTVARLKA GEIIVRKGET GDEVERFFLS DGFVLFKSPE DDSGCCTAEV LGVEVVPVSM LDKESAATAL QELLQQGAGA T DEWTKART LLGQELLSSV IRAAP |
-Macromolecule #5: ATP synthase subunit epsilon
| Macromolecule | Name: ATP synthase subunit epsilon / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Toxoplasma gondii (strain ATCC 50853 / GT1) (eukaryote) Toxoplasma gondii (strain ATCC 50853 / GT1) (eukaryote)Strain: ATCC 50853 / GT1 |
| Molecular weight | Theoretical: 8.492709 KDa |
| Sequence | String: MWRSSGVSFT RYASEMAALL RQCLKEPYRT QAMQRNQIHL KETVYQQGQV LTRETFNDIK KAFEAAAKHA GEK |
-Macromolecule #6: subunit c
| Macromolecule | Name: subunit c / type: protein_or_peptide / ID: 6 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Toxoplasma gondii (strain ATCC 50853 / GT1) (eukaryote) Toxoplasma gondii (strain ATCC 50853 / GT1) (eukaryote)Strain: ATCC 50853 / GT1 |
| Molecular weight | Theoretical: 17.753504 KDa |
| Sequence | String: MFFSRLSLSA LKAAPAREAL PGLLSRQSFS SAGFSQFSSQ KFFFSPSRNF SQSPLFQKHT PVHCNQRIAS ALVPTQQPAM TRQNPYAMQ VGARYDAGVA SLSAAIALMS VGGVAQGIGS LFAALVSGTA RNPSIKEDLF TYTLIGMGFL EFLGIICVLM S AVLLYS |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV / Details: 3 seconds blot.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal magnification: 165000 Bright-field microscopy / Cs: 2.7 mm / Nominal magnification: 165000 |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Number real images: 4860 / Average electron dose: 30.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-6tmj: |
 Movie
Movie Controller
Controller



















 Z
Z Y
Y X
X