+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10313 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
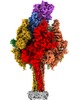
| Title | Structure of Photorhabdus luminescens Tc holotoxin pore | |||||||||
 Map data Map data | Half map 1 | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |   Photorhabdus luminescens (bacteria) Photorhabdus luminescens (bacteria) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.4 Å cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Roderer D / Raunser S | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2019 Journal: Proc Natl Acad Sci U S A / Year: 2019Title: Structure of a Tc holotoxin pore provides insights into the translocation mechanism. Authors: Daniel Roderer / Oliver Hofnagel / Roland Benz / Stefan Raunser /  Abstract: Tc toxins are modular toxin systems of insect and human pathogenic bacteria. They are composed of a 1.4-MDa pentameric membrane translocator (TcA) and a 250-kDa cocoon (TcB and TcC) encapsulating the ...Tc toxins are modular toxin systems of insect and human pathogenic bacteria. They are composed of a 1.4-MDa pentameric membrane translocator (TcA) and a 250-kDa cocoon (TcB and TcC) encapsulating the 30-kDa toxic enzyme (C terminus of TcC). Binding of Tc toxins to target cells and a pH shift trigger the conformational transition from the soluble prepore state to the membrane-embedded pore. Subsequently, the toxic enzyme is translocated and released into the cytoplasm. A high-resolution structure of a holotoxin embedded in membranes is missing, leaving open the question of whether TcB-TcC has an influence on the conformational transition of TcA. Here we show in atomic detail a fully assembled 1.7-MDa Tc holotoxin complex from in the membrane. We find that the 5 TcA protomers conformationally adapt to fit around the cocoon during the prepore-to-pore transition. The architecture of the Tc toxin complex allows TcB-TcC to bind to an already membrane-embedded TcA pore to form a holotoxin. Importantly, assembly of the holotoxin at the membrane results in spontaneous translocation of the toxic enzyme, indicating that this process is not driven by a proton gradient or other energy source. Mammalian lipids with zwitterionic head groups are preferred over other lipids for the integration of Tc toxins. In a nontoxic Tc toxin variant, we can visualize part of the translocating toxic enzyme, which transiently interacts with alternating negative charges and hydrophobic stretches of the translocation channel, providing insights into the mechanism of action of Tc toxins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10313.map.gz emd_10313.map.gz | 17.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10313-v30.xml emd-10313-v30.xml emd-10313.xml emd-10313.xml | 15.4 KB 15.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10313.png emd_10313.png | 122.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10313 http://ftp.pdbj.org/pub/emdb/structures/EMD-10313 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10313 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10313 | HTTPS FTP |
-Related structure data
| Related structure data |  6sufMC  6sueC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10313.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10313.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Tc holotoxin complex in the pore form, formed by the TcdA1 pentam...
| Entire | Name: Tc holotoxin complex in the pore form, formed by the TcdA1 pentamer and TcdB2-TccC3 |
|---|---|
| Components |
|
-Supramolecule #1: Tc holotoxin complex in the pore form, formed by the TcdA1 pentam...
| Supramolecule | Name: Tc holotoxin complex in the pore form, formed by the TcdA1 pentamer and TcdB2-TccC3 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Photorhabdus luminescens (bacteria) Photorhabdus luminescens (bacteria) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 1.7 MDa |
-Macromolecule #1: TcdA1
| Macromolecule | Name: TcdA1 / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Photorhabdus luminescens (bacteria) Photorhabdus luminescens (bacteria) |
| Molecular weight | Theoretical: 283.230375 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MNESVKEIPD VLKSQCGFNC LTDISHSSFN EFRQQVSEHL SWSETHDLYH DAQQAQKDNR LYEARILKRA NPQLQNAVHL AILAPNAEL IGYNNQFSGR ASQYVAPGTV SSMFSPAAYL TELYREARNL HASDSVYYLD TRRPDLKSMA LSQQNMDIEL S TLSLSNEL ...String: MNESVKEIPD VLKSQCGFNC LTDISHSSFN EFRQQVSEHL SWSETHDLYH DAQQAQKDNR LYEARILKRA NPQLQNAVHL AILAPNAEL IGYNNQFSGR ASQYVAPGTV SSMFSPAAYL TELYREARNL HASDSVYYLD TRRPDLKSMA LSQQNMDIEL S TLSLSNEL LLESIKTESK LENYTKVMEM LSTFRPSGAT PYHDAYENVR EVIQLQDPGL EQLNASPAIA GLMHQASLLG IN ASISPEL FNILTEEITE GNAEELYKKN FGNIEPASLA MPEYLKRYYN LSDEELSQFI GKASNFGQQE YSNNQLITPV VNS SDGTVK VYRITREYTT NAYQMDVELF PFGGENYRLD YKFKNFYNAS YLSIKLNDKR ELVRTEGAPQ VNIEYSANIT LNTA DISQP FEIGLTRVLP SGSWAYAAAK FTVEEYNQYS FLLKLNKAIR LSRATELSPT ILEGIVRSVN LQLDINTDVL GKVFL TKYY MQRYAIHAET ALILCNAPIS QRSYDNQPSQ FDRLFNTPLL NGQYFSTGDE EIDLNSGSTG DWRKTILKRA FNIDDV SLF RLLKITDHDN KDGKIKNNLK NLSNLYIGKL LADIHQLTID ELDLLLIAVG EGKTNLSAIS DKQLATLIRK LNTITSW LH TQKWSVFQLF IMTSTSYNKT LTPEIKNLLD TVYHGLQGFD KDKADLLHVM APYIAATLQL SSENVAHSVL LWADKLQP G DGAMTAEKFW DWLNTKYTPG SSEAVETQEH IVQYCQALAQ LEMVYHSTGI NENAFRLFVT KPEMFGAATG AAPAHDALS LIMLTRFADW VNALGEKASS VLAAFEANSL TAEQLADAMN LDANLLLQAS IQAQNHQHLP PVTPENAFSC WTSINTILQW VNVAQQLNV APQGVSALVG LDYIESMKET PTYAQWENAA GVLTAGLNSQ QANTLHAFLD ESRSAALSTY YIRQVAKAAA A IKSRDDLY QYLLIDNQVS AAIKTTRIAE AIASIQLYVN RALENVEENA NSGVISRQFF IDWDKYNKRY STWAGVSQLV YY PENYIDP TMRIGQTKMM DALLQSVSQS QLNADTVEDA FMSYLTSFEQ VANLKVISAY HDNINNDQGL TYFIGLSETD AGE YYWRSV DHSKFNDGKF AANAWSEWHK IDCPINPYKS TIRPVIYKSR LYLLWLEQKE ITKQTGNSKD GYQTETDYRY ELKL AHIRY DGTWNTPITF DVNKKISELK LEKNRAPGLY CAGYQGEDTL LVMFYNQQDT LDSYKNASMQ GLYIFADMAS KDMTP EQSN VYRDNSYQQF DTNNVRRVNN RYAEDYEIPS SVSSRKDYGW GDYYLSMVYN GDIPTINYKA ASSDLKIYIS PKLRII HNG YEGQKRNQCN LMNKYGKLGD KFIVYTSLGV NPNNSSNKLM FYPVYQYSGN TSGLNQGRLL FHRDTTYPSK VEAWIPG AK RSLTNQNAAI GDDYATDSLN KPDDLKQYIF MTDSKGTATD VSGPVEINTA ISPAKVQIIV KAGGKEQTFT ADKDVSIQ P SPSFDEMNYQ FNALEIDGSG LNFINNSASI DVTFTAFAED GRKLGYESFS IPVTLKVSTD NALTLHHNEN GAQYMQWQS YRTRLNTLFA RQLVARATTG IDTILSMETQ NIQEPQLGKG FYATFVIPPY NLSTHGDERW FKLYIKHVVD NNSHIIYSGQ LTDTNINIT LFIPLDDVPL NQDYHAKVYM TFKKSPSDGT WWGPHFVRDD KGIVTINPKS ILTHFESVNV LNNISSEPMD F SGANSLYF WELFYYTPML VAQRLLHEQN FDEANRWLKY VWSPSGYIVH GQIQNYQWNV RPLLEDTSWN SDPLDSVDPD AV AQHDPMH YKVSTFMRTL DLLIARGDHA YRQLERDTLN EAKMWYMQAL HLLGDKPYLP LSTTWSDPRL DRAADITTQN AHD SAIVAL RQNIPTPAPL SLRSANTLTD LFLPQINEVM MNYWQTLAQR VYNLRHNLSI DGQPLYLPIY ATPADPKALL SAAV ATSQG GGKLPESFMS LWRFPHMLEN ARGMVSQLTQ FGSTLQNIIE RQDAEALNAL LQNQAAELIL TNLSIQDKTI EELDA EKTV LEKSKAGAQS RFDSYGKLYD ENINAGENQA MTLRASAAGL TTAVQASRLA GAAADLVPNI FGFAGGGSRW GAIAEA TGY VMEFSANVMN TEADKISQSE TYRRRRQEWE IQRNNAEAEL KQIDAQLKSL AVRREAAVLQ KTSLKTQQEQ TQSQLAF LQ RKFSNQALYN WLRGRLAAIY FQFYDLAVAR CLMAEQAYRW ELNDDSARFI KPGAWQGTYA GLLAGETLML SLAQMEDA H LKRDKRALEV ERTVSLAEVY AGLPKDNGPF SLAQEIDKLV SQGSGSAGSG NNNLAFGAGT DTKTSLQASV SFADLKIRE DYPASLGKIR RIKQISVTLP ALLGPYQDVQ AILSYGDKAG LANGCEALAV SHGMNDSGQF QLDFNDGKFL PFEGIAIDQG TLTLSFPNA SMPEKGKQAT MLKTLNDIIL HIRYTIK |
-Macromolecule #2: TcdB2,TccC3
| Macromolecule | Name: TcdB2,TccC3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Photorhabdus luminescens (bacteria) Photorhabdus luminescens (bacteria) |
| Molecular weight | Theoretical: 274.18825 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MQNSQDFSIT ELSLPKGGGA ITGMGEALTP TGPDGMAALS LPLPISAGRG YAPAFTLNYN SGAGNSPFGL GWDCNVMTIR RRTHFGVPH YDETDTFLGP EGEVLVVADQ PRDESTLQGI NLGATFTVTG YRSRLESHFS RLEYWQPKTT GKTDFWLIYS P DGQVHLLG ...String: MQNSQDFSIT ELSLPKGGGA ITGMGEALTP TGPDGMAALS LPLPISAGRG YAPAFTLNYN SGAGNSPFGL GWDCNVMTIR RRTHFGVPH YDETDTFLGP EGEVLVVADQ PRDESTLQGI NLGATFTVTG YRSRLESHFS RLEYWQPKTT GKTDFWLIYS P DGQVHLLG KSPQARISNP SQTTQTAQWL LEASVSSRGE QIYYQYRAED DTGCEADEIT HHLQATAQRY LHIVYYGNRT AS ETLPGLD GSAPSQADWL FYLVFDYGER SNNLKTPPAF STTGSWLCRQ DRFSRYEYGF EIRTRRLCRQ VLMYHHLQAL DSK ITEHNG PTLVSRLILN YDESAIASTL VFVRRVGHEQ DGNVVTLPPL ELAYQDFSPR HHAHWQPMDV LANFNAIQRW QLVD LKGEG LPGLLYQDKG AWWYRSAQRL GEIGSDAVTW EKMQPLSVIP SLQSNASLVD INGDGQLDWV ITGPGLRGYH SQRPD GSWT RFTPLNALPV EYTHPRAQLA DLMGAGLSDL VLIGPKSVRL YANTRDGFAK GKDVVQSGEI TLPVPGADPR KLVAFS DVL GSGQAHLVEV SATKVTCWPN LGRGRFGQPI TLPGFSQPAT EFNPAQVYLA DLDGSGPTDL IYVHTNRLDI FLNKSGN GF AEPVTLRFPE GLRFDHTCQL QMADVQGLGV ASLILSVPHM SPHHWRCDLT NMKPWLLNEM NNNMGVHHTL RYRSSSQF W LDEKAAALTT GQTPVCYLPF PIHTLWQTET EDEISGNKLV TTLRYARGAW DGREREFRGF GYVEQTDSHQ LAQGNAPER TPPALTKNWY ATGLPVIDNA LSTEYWRDDQ AFAGFSPRFT TWQDNKDVPL TPEDDNSRYW FNRALKGQLL RSELYGLDDS TNKHVPYTV TEFRSQVRRL QHTDSRYPVL WSSVVESRNY HYERIASDPQ CSQNITLSSD RFGQPLKQLS VQYPRRQQPA I NLYPDTLP DKLLANSYDD QQRQLRLTYQ QSSWHHLTNN TVRVLGLPDS TRSDIFTYGA ENVPAGGLNL ELLSDKNSLI AD DKPREYL GQQKTAYTDG QNTTPLQTPT RQALIAFTET TVFNQSTLSA FNGSIPSDKL STTLEQAGYQ QTNYLFPRTG EDK VWVAHH GYTDYGTAAQ FWRPQKQSNT QLTGKITLIW DANYCVVVQT RDAAGLTTSA KYDWRFLTPV QLTDINDNQH LITL DALGR PITLRFWGTE NGKMTGYSSP EKASFSPPSD VNAAIELKKP LPVAQCQVYA PESWMPVLSQ KTFNRLAEQD WQKLY NARI ITEDGRICTL AYRRWVQSQK AIPQLISLLN NGPRLPPHSL TLTTDRYDHD PEQQIRQQVV FSDGFGRLLQ AAARHE AGM ARQRNEDGSL IINVQHTENR WAVTGRTEYD NKGQPIRTYQ PYFLNDWRYV SNDSARQEKE AYADTHVYDP IGREIKV IT AKGWFRRTLF TPWFTVNEDE NDTAAEVKKV KMPGSRPMKN IDPKLYQKTP TVSVYDNRGL IIRNIDFHRT TANGDPDT R ITRHQYDIHG HLNQSIDPRL YEAKQTNNTI KPNFLWQYDL TGNPLCTESI DAGRTVTLND IEGRPLLTVT ATGVIQTRQ YETSSLPGRL LSVAEQTPEE KTSRITERLI WAGNTEAEKD HNLAGQCVRH YDTAGVTRLE SLSLTGTVLS QSSQLLIDTQ EANWTGDNE TVWQNMLADD IYTTLSTFDA TGALLTQTDA KGNIQRLAYD VAGQLNGSWL TLKGQTEQVI IKSLTYSAAG Q KLREEHGN DVITEYSYEP ETQRLIGIKT RRPSDTKVLQ DLRYEYDPVG NVISIRNDAE ATRFWHNQKV MPENTYTYDS LY QLISATG REMANIGQQS HQFPSPALPS DNNTYTNYTR TYTYDRGGNL TKIQHSSPAT QNNYTTNITV SNRSNRAVLS TLT EDPAQV DALFDAGGHQ NTLISGQNLN WNTRGELQQV TLVKRDKGAN DDREWYRYSG DGRRMLKINE QQASNNAQTQ RVTY LPNLE LRLTQNSTAT TEDLQVITVG EAGRAQVRVL HWESGKPEDI DNNQLRYSYD NLIGSSQLEL DSEGQIISEE EYYPY GGTA LWAARNQTEA SYKTIRYSGK ERDATGLYYY GYRYYQPWIG RWLSSDPAGT IDGLNLYRMV RNNPVTLLDP DGLMPT IAE RIAALKKNKV TDSAPSPANA TNVAINIRPP VAPKPSLPKA STSSQPTTHP IGAANIKPTT SGSSIVAPLS PVGNKST SE ISLPESAQSS SSSTTSTNLQ KKSFTLYRAD NRSFEEMQSK FPEGFKAWTP LDTKMARQFA SIFIGQKDTS NLPKETVK N ISTWGAKPKL KDLSNYIKYT KDKSTVWVST AINTEAGGQS SGAPLHKIDM DLYEFAIDGQ KLNPLPEGRT KNMVPSLLL DTPQIETSSI IALNHGPVND AEISFLTTIP LKNVKPHKR |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 60.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Initial angle assignment | Type: NOT APPLICABLE |
|---|---|
| Final angle assignment | Type: NOT APPLICABLE |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: SPHIRE / Number images used: 64806 |
 Movie
Movie Controller
Controller