[English] 日本語
 Yorodumi
Yorodumi- EMDB-10134: CryoEM structure of murine perforin-2 ectodomain in a pre-pore form -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10134 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of murine perforin-2 ectodomain in a pre-pore form | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationdendritic cell antigen processing and presentation / antigen processing and presentation of exogenous peptide antigen / phagolysosome membrane /  endolysosome / pore-forming activity / antibacterial innate immune response / wide pore channel activity / antigen processing and presentation of exogenous peptide antigen via MHC class I / phagocytic vesicle / phagocytic vesicle membrane ...dendritic cell antigen processing and presentation / antigen processing and presentation of exogenous peptide antigen / phagolysosome membrane / endolysosome / pore-forming activity / antibacterial innate immune response / wide pore channel activity / antigen processing and presentation of exogenous peptide antigen via MHC class I / phagocytic vesicle / phagocytic vesicle membrane ...dendritic cell antigen processing and presentation / antigen processing and presentation of exogenous peptide antigen / phagolysosome membrane /  endolysosome / pore-forming activity / antibacterial innate immune response / wide pore channel activity / antigen processing and presentation of exogenous peptide antigen via MHC class I / phagocytic vesicle / phagocytic vesicle membrane / cytoplasmic vesicle / defense response to Gram-negative bacterium / endolysosome / pore-forming activity / antibacterial innate immune response / wide pore channel activity / antigen processing and presentation of exogenous peptide antigen via MHC class I / phagocytic vesicle / phagocytic vesicle membrane / cytoplasmic vesicle / defense response to Gram-negative bacterium /  adaptive immune response / defense response to Gram-positive bacterium / defense response to bacterium adaptive immune response / defense response to Gram-positive bacterium / defense response to bacteriumSimilarity search - Function | |||||||||
| Biological species |   Mus musculus (house mouse) Mus musculus (house mouse) | |||||||||
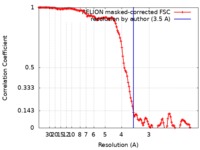
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.5 Å cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Ni T / Yu X / Gilbert RJC | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2020 Journal: Sci Adv / Year: 2020Title: Structure and mechanism of bactericidal mammalian perforin-2, an ancient agent of innate immunity. Authors: Tao Ni / Fang Jiao / Xiulian Yu / Saša Aden / Lucy Ginger / Sophie I Williams / Fangfang Bai / Vojtěch Pražák / Dimple Karia / Phillip Stansfeld / Peijun Zhang / George Munson / Gregor ...Authors: Tao Ni / Fang Jiao / Xiulian Yu / Saša Aden / Lucy Ginger / Sophie I Williams / Fangfang Bai / Vojtěch Pražák / Dimple Karia / Phillip Stansfeld / Peijun Zhang / George Munson / Gregor Anderluh / Simon Scheuring / Robert J C Gilbert /    Abstract: Perforin-2 (MPEG1) is thought to enable the killing of invading microbes engulfed by macrophages and other phagocytes, forming pores in their membranes. Loss of perforin-2 renders individual ...Perforin-2 (MPEG1) is thought to enable the killing of invading microbes engulfed by macrophages and other phagocytes, forming pores in their membranes. Loss of perforin-2 renders individual phagocytes and whole organisms significantly more susceptible to bacterial pathogens. Here, we reveal the mechanism of perforin-2 activation and activity using atomic structures of pre-pore and pore assemblies, high-speed atomic force microscopy, and functional assays. Perforin-2 forms a pre-pore assembly in which its pore-forming domain points in the opposite direction to its membrane-targeting domain. Acidification then triggers pore formation, via a 180° conformational change. This novel and unexpected mechanism prevents premature bactericidal attack and may have played a key role in the evolution of all perforin family proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10134.map.gz emd_10134.map.gz | 14.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10134-v30.xml emd-10134-v30.xml emd-10134.xml emd-10134.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10134_fsc.xml emd_10134_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_10134.png emd_10134.png | 80.3 KB | ||
| Masks |  emd_10134_msk_1.map emd_10134_msk_1.map | 178 MB |  Mask map Mask map | |
| Others |  emd_10134_half_map_1.map.gz emd_10134_half_map_1.map.gz emd_10134_half_map_2.map.gz emd_10134_half_map_2.map.gz | 135.6 MB 135.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10134 http://ftp.pdbj.org/pub/emdb/structures/EMD-10134 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10134 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10134 | HTTPS FTP |
-Related structure data
| Related structure data |  6sb3MC  6sb1C  6sb4C  6sb5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10134.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10134.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10134_msk_1.map emd_10134_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
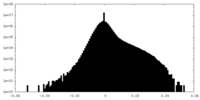
| Density Histograms |
-Half map: #1
| File | emd_10134_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
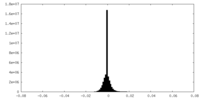
| Density Histograms |
-Half map: #2
| File | emd_10134_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
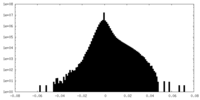
| Density Histograms |
- Sample components
Sample components
-Entire : Murine perforin-2 ecto domain
| Entire | Name: Murine perforin-2 ecto domain |
|---|---|
| Components |
|
-Supramolecule #1: Murine perforin-2 ecto domain
| Supramolecule | Name: Murine perforin-2 ecto domain / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant cell: HEK293T / Recombinant plasmid: pHLsec-1D4 Homo sapiens (human) / Recombinant cell: HEK293T / Recombinant plasmid: pHLsec-1D4 |
-Macromolecule #1: Macrophage-expressed gene 1 protein
| Macromolecule | Name: Macrophage-expressed gene 1 protein / type: protein_or_peptide / ID: 1 / Number of copies: 16 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 71.129531 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ETGDKPLGET GTTGFQICKN ALKLPVLEVL PGGGWDNLRN VDMGRVMDLT YTNCKTTEDG QYIIPDEVYT IPQKESNLEM NSEVLESWM NYQSTTSLSI NTELALFSRV NGKFSTEFQR MKTLQVKDQA VTTRVQVRNR IYTVKTTPTS ELSLGFTKAL M DICDQLEK ...String: ETGDKPLGET GTTGFQICKN ALKLPVLEVL PGGGWDNLRN VDMGRVMDLT YTNCKTTEDG QYIIPDEVYT IPQKESNLEM NSEVLESWM NYQSTTSLSI NTELALFSRV NGKFSTEFQR MKTLQVKDQA VTTRVQVRNR IYTVKTTPTS ELSLGFTKAL M DICDQLEK NQTKMATYLA ELLILNYGTH VITSVDAGAA LVQEDHVRSS FLLDNQNSQN TVTASAGIAF LNIVNFKVET DY ISQTSLT KDYLSNRTNS RVQSFGGVPF YPGITLETWQ KGITNHLVAI DRAGLPLHFF IKPDKLPGLP GPLVKKLSKT VET AVRHYY TFNTHPGCTN VDSPNFNFQA NMDDDSCDAK VTNFTFGGVY QECTELSGDV LCQNLEQKNL LTGDFSCPPG YSPV HLLSQ THEEGYSRLE CKKKCTLKIF CKTVCEDVFR VAKAEFRAYW CVAAGQVPDN SGLLFGGVFT DKTINPMTNA QSCPA GYIP LNLFESLKVC VSLDYELGFK FSVPFGGFFS CIMGNPLVNS DTAKDVRAPS LKKCPGGFSQ HLAVISDGCQ VSYCVK AGI FTGGSLLPVR LPPYTKPPLM SQVATNTVIV TNSETARSWI KDPQTNQWKL GEPLELRRAM TVIHGDSNGM SGGGTET SQ VAPA |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 32 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 5.5 |
|---|---|
| Grid | Model: Homemade / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z
Z Y
Y X
X