[English] 日本語
 Yorodumi
Yorodumi- SASDHC2: PieE-FAD: 2,4-dichlorophenol 6-monooxygenase bound to flavin aden... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 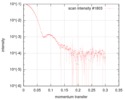 |
|---|---|
 Sample Sample | PieE-FAD: 2,4-dichlorophenol 6-monooxygenase bound to flavin adenine dinucleotide
|
| Function / homology | Aromatic-ring hydroxylase, C-terminal / FAD-binding domain / FAD binding domain / FAD binding /  monooxygenase activity / FAD/NAD(P)-binding domain superfamily / monooxygenase activity / FAD/NAD(P)-binding domain superfamily /  2,4-dichlorophenol 6-monooxygenase 2,4-dichlorophenol 6-monooxygenase Function and homology information Function and homology information |
| Biological species |  Streptomyces sp. SCSIO 03032 (bacteria) Streptomyces sp. SCSIO 03032 (bacteria) |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDHC2 SASDHC2 |
|---|
-Related structure data
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
|---|
-Models
- Sample
Sample
 Sample Sample | Name: PieE-FAD: 2,4-dichlorophenol 6-monooxygenase bound to flavin adenine dinucleotide Specimen concentration: 17 mg/ml / Entity id: 1962 / 1963 |
|---|---|
| Buffer | Name: 20 mM Tris, 150 mM NaCl, 5 mM DTT, 2% glycerol / pH: 7.5 |
| Entity #1962 | Name: PieE / Type: protein / Description: 2,4-dichlorophenol 6-monooxygenase / Formula weight: 66.552 / Num. of mol.: 6 / Source: Streptomyces sp. SCSIO 03032 / References: UniProt: W0C4C9 / Formula weight: 66.552 / Num. of mol.: 6 / Source: Streptomyces sp. SCSIO 03032 / References: UniProt: W0C4C9Sequence: MGSSHHHHHH SSGLVPRGSH MTGSEGTAPD IRVPVLIVGG GPAGLTAALA LSRYGVPHLL VNRHHGTAHT PRAHLLNQRT GEIFRDLGIA DRVEAHATPG HLMANHVFMS TFAGPEVARI GAYGNGPDRI GEYRAASPSG LCNLPQHLLE PLLVEAVQEA CVGQLRFGHE ...Sequence: MGSSHHHHHH SSGLVPRGSH MTGSEGTAPD IRVPVLIVGG GPAGLTAALA LSRYGVPHLL VNRHHGTAHT PRAHLLNQRT GEIFRDLGIA DRVEAHATPG HLMANHVFMS TFAGPEVARI GAYGNGPDRI GEYRAASPSG LCNLPQHLLE PLLVEAVQEA CVGQLRFGHE FVSLEQDEHG VTSRITDRRT GRDYTVRSDY LIGADGARSR VLAQLGIALD GATGIARAVT TWFEADLSRY SAHRPALLYM GAVPGSPPAD GRVFVSLRPW TEWLHLTFPP PTADVDVEDH EAVRAGIRES IGDPTVDVTI KNVSAWEVNS AVAPRYASGR VFCVGDAVHQ NPPTNGLGLN SAVADSFNLC WKLKLALEGL AGPGLLDTYH DERQPVGRQI VDRAFRSMVD LIGIPQALGF TEGQSPEEQW RLLDTLHEDT EEARQRRAAL AAATAAIHGQ ANAHGVELGY RYRTGALVPD GTPEPADERD PELYYRATTW PGARLPHAWL ENGRHRCSTL DVTGRGRFTL LTGPGGEPWR DAARDAALDT GVEVAVLPIG AGGGPRDPYG TWAELREVEE SGAVLVRPDG HVAWRARDHG HAKELPEVMA RVLHQPDPAA RRTGGPGA |
| Entity #1963 | Name: FAD / Type: other / Description: Flavin adenine dinucleotide / Formula weight: 0.786 / Num. of mol.: 6 / Formula weight: 0.786 / Num. of mol.: 6 Sequence: C27H33N9O1 5P2 |
-Experimental information
| Beam | Instrument name: Université de Montréal Xenocs BioXolver L with MetalJet City: Montréal / 国: Canada  / Type of source: X-ray in house / Type of source: X-ray in house / Wavelength: 0.134 Å / Dist. spec. to detc.: 0.6 mm / Wavelength: 0.134 Å / Dist. spec. to detc.: 0.6 mm | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus3 R 300K / Type: pixel counting / Pixsize x: 172 mm | |||||||||||||||||||||||||||||||||
| Scan | Measurement date: Oct 22, 2019 / Storage temperature: 24 °C / Cell temperature: 20 °C / Exposure time: 30 sec. / Number of frames: 190 / Unit: 1/A /
| |||||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| |||||||||||||||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller


