[English] 日本語
 Yorodumi
Yorodumi- SASDFU3: Lysine-specific Demethylase (LSD2) (Lysyne-specific Demethylase L... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 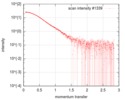 |
|---|---|
 Sample Sample | Lysine-specific Demethylase (LSD2)
|
| Function / homology |  Function and homology information Function and homology informationepigenetic programing of female pronucleus / [histone H3]-N6,N6-dimethyl-L-lysine4 FAD-dependent demethylase / FAD-dependent H3K4me/H3K4me3 demethylase activity /  genomic imprinting / histone demethylase activity / transcription initiation-coupled chromatin remodeling / NR1H3 & NR1H2 regulate gene expression linked to cholesterol transport and efflux / FAD binding / HDMs demethylate histones / UCH proteinases ...epigenetic programing of female pronucleus / [histone H3]-N6,N6-dimethyl-L-lysine4 FAD-dependent demethylase / FAD-dependent H3K4me/H3K4me3 demethylase activity / genomic imprinting / histone demethylase activity / transcription initiation-coupled chromatin remodeling / NR1H3 & NR1H2 regulate gene expression linked to cholesterol transport and efflux / FAD binding / HDMs demethylate histones / UCH proteinases ...epigenetic programing of female pronucleus / [histone H3]-N6,N6-dimethyl-L-lysine4 FAD-dependent demethylase / FAD-dependent H3K4me/H3K4me3 demethylase activity /  genomic imprinting / histone demethylase activity / transcription initiation-coupled chromatin remodeling / NR1H3 & NR1H2 regulate gene expression linked to cholesterol transport and efflux / FAD binding / HDMs demethylate histones / UCH proteinases / genomic imprinting / histone demethylase activity / transcription initiation-coupled chromatin remodeling / NR1H3 & NR1H2 regulate gene expression linked to cholesterol transport and efflux / FAD binding / HDMs demethylate histones / UCH proteinases /  nucleosome / nucleosome /  flavin adenine dinucleotide binding / flavin adenine dinucleotide binding /  histone binding / histone binding /  oxidoreductase activity / oxidoreductase activity /  chromatin binding / chromatin binding /  chromatin / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / zinc ion binding / chromatin / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / zinc ion binding /  nucleoplasm / nucleoplasm /  nucleus nucleusSimilarity search - Function |
| Biological species |   Homo sapiens (human) Homo sapiens (human) |
 Citation Citation |  Journal: Cell Rep / Year: 2019 Journal: Cell Rep / Year: 2019Title: A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex. Authors: Chiara Marabelli / Biagina Marrocco / Simona Pilotto / Sagar Chittori / Sarah Picaud / Sara Marchese / Giuseppe Ciossani / Federico Forneris / Panagis Filippakopoulos / Guy Schoehn / Daniela ...Authors: Chiara Marabelli / Biagina Marrocco / Simona Pilotto / Sagar Chittori / Sarah Picaud / Sara Marchese / Giuseppe Ciossani / Federico Forneris / Panagis Filippakopoulos / Guy Schoehn / Daniela Rhodes / Sriram Subramaniam / Andrea Mattevi /       Abstract: LSD1 and LSD2 are homologous histone demethylases with opposite biological outcomes related to chromatin silencing and transcription elongation, respectively. Unlike LSD1, LSD2 nucleosome-demethylase ...LSD1 and LSD2 are homologous histone demethylases with opposite biological outcomes related to chromatin silencing and transcription elongation, respectively. Unlike LSD1, LSD2 nucleosome-demethylase activity relies on a specific linker peptide from the multidomain protein NPAC. We used single-particle cryoelectron microscopy (cryo-EM), in combination with kinetic and mutational analysis, to analyze the mechanisms underlying the function of the human LSD2/NPAC-linker/nucleosome complex. Weak interactions between LSD2 and DNA enable multiple binding modes for the association of the demethylase to the nucleosome. The demethylase thereby captures mono- and dimethyl Lys4 of the H3 tail to afford histone demethylation. Our studies also establish that the dehydrogenase domain of NPAC serves as a catalytically inert oligomerization module. While LSD1/CoREST forms a nucleosome docking platform at silenced gene promoters, LSD2/NPAC is a multifunctional enzyme complex with flexible linkers, tailored for rapid chromatin modification, in conjunction with the advance of the RNA polymerase on actively transcribed genes. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDFU3 SASDFU3 |
|---|
-Related structure data
| Related structure data |  4704C  4705C  4710C  4711C  4712C  6r1tC  6r1uC  6r25C C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
- Sample
Sample
 Sample Sample | Name: Lysine-specific Demethylase (LSD2) / Specimen concentration: 2 mg/ml |
|---|---|
| Buffer | Name: 15 mM HEPES, 200 mM NaCl / pH: 7.3 |
| Entity #1516 | Name: LSD2 / Type: protein / Description: Lysyne-specific Demethylase LSD2 / Formula weight: 88.832 / Num. of mol.: 1 / Source: Homo sapiens / References: UniProt: Q8NB78 Sequence: KKKATETTDE DEDGGSEKKY RKCEKAGCTA TCPVCFASAS ERCAKNGYTS RWYHLSCGEH FCNECFDHYY RSHKDGYDKY TTWKKIWTSN GKTEPSPKAF MADQQLPYWV QCTKPECRKW RQLTKEIQLT PQIAKTYRCG MKPNTAIKPE TSDHCSLPED LRVLEVSNHW ...Sequence: KKKATETTDE DEDGGSEKKY RKCEKAGCTA TCPVCFASAS ERCAKNGYTS RWYHLSCGEH FCNECFDHYY RSHKDGYDKY TTWKKIWTSN GKTEPSPKAF MADQQLPYWV QCTKPECRKW RQLTKEIQLT PQIAKTYRCG MKPNTAIKPE TSDHCSLPED LRVLEVSNHW WYSMLILPPL LKDSVAAPLL SAYYPDCVGM SPSCTSTNRA AATGNASPGK LEHSKAALSV HVPGMNRYFQ PFYQPNECGK ALCVRPDVME LDELYEFPEY SRDPTMYLAL RNLILALWYT NCKEALTPQK CIPHIIVRGL VRIRCVQEVE RILYFMTRKG LINTGVLSVG ADQYLLPKDY HNKSVIIIGA GPAGLAAARQ LHNFGIKVTV LEAKDRIGGR VWDDKSFKGV TVGRGAQIVN GCINNPVALM CEQLGISMHK FGERCDLIQE GGRITDPTID KRMDFHFNAL LDVVSEWRKD KTQLQDVPLG EKIEEIYKAF IKESGIQFSE LEGQVLQFHL SNLEYACGSN LHQVSARSWD HNEFFAQFAG DHTLLTPGYS VIIEKLAEGL DIQLKSPVQC IDYSGDEVQV TTTDGTGYSA QKVLVTVPLA LLQKGAIQFN PPLSEKKMKA INSLGAGIIE KIALQFPYRF WDSKVQGADF FGHVPPSASK RGLFAVFYDM DPQKKHSVLM SVIAGEAVAS VRTLDDKQVL QQCMATLREL FKEQEVPDPT KYFVTRWSTD PWIQMAYSFV KTGGSGEAYD IIAEDIQGTV FFAGEATNRH FPQTVTGAYL SGVREASKIA AF |
-Experimental information
| Beam | Instrument name: ESRF BM29 / City: Grenoble / 国: France  / Type of source: X-ray synchrotron / Type of source: X-ray synchrotron Synchrotron / Wavelength: 0.1 Å / Dist. spec. to detc.: 2.87 mm Synchrotron / Wavelength: 0.1 Å / Dist. spec. to detc.: 2.87 mm | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 1M / Type: Dectris / Pixsize x: 172 mm | ||||||||||||||||||||||||||||||
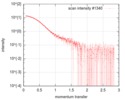
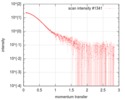
| Scan |
| ||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| ||||||||||||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller