[English] 日本語
 Yorodumi
Yorodumi- SASDFD2: wild-type human Latent Transforming Growth Factor beta 1 (LTGFB-1) -
+ Open data
Open data
- Basic information
Basic information
| Entry | 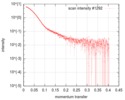 |
|---|---|
 Sample Sample | wild-type human Latent Transforming Growth Factor beta 1 (LTGFB-1)
|
| Function / homology |  Function and homology information Function and homology informationpositive regulation of primary miRNA processing / positive regulation of microglia differentiation / Influenza Virus Induced Apoptosis / negative regulation of skeletal muscle tissue development / TGFBR2 MSI Frameshift Mutants in Cancer / regulatory T cell differentiation / regulation of blood vessel remodeling / regulation of striated muscle tissue development / negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target / regulation of protein import into nucleus ...positive regulation of primary miRNA processing / positive regulation of microglia differentiation / Influenza Virus Induced Apoptosis / negative regulation of skeletal muscle tissue development / TGFBR2 MSI Frameshift Mutants in Cancer / regulatory T cell differentiation / regulation of blood vessel remodeling / regulation of striated muscle tissue development / negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target / regulation of protein import into nucleus /  extracellular matrix assembly / embryonic liver development / type III transforming growth factor beta receptor binding / negative regulation of hyaluronan biosynthetic process / positive regulation of cardiac muscle cell differentiation / myofibroblast differentiation / connective tissue replacement involved in inflammatory response wound healing / odontoblast differentiation / negative regulation of macrophage cytokine production / positive regulation of receptor signaling pathway via STAT / TGFBR2 Kinase Domain Mutants in Cancer / positive regulation of isotype switching to IgA isotypes / positive regulation of mesenchymal stem cell proliferation / extracellular matrix assembly / embryonic liver development / type III transforming growth factor beta receptor binding / negative regulation of hyaluronan biosynthetic process / positive regulation of cardiac muscle cell differentiation / myofibroblast differentiation / connective tissue replacement involved in inflammatory response wound healing / odontoblast differentiation / negative regulation of macrophage cytokine production / positive regulation of receptor signaling pathway via STAT / TGFBR2 Kinase Domain Mutants in Cancer / positive regulation of isotype switching to IgA isotypes / positive regulation of mesenchymal stem cell proliferation /  membrane protein intracellular domain proteolysis / SMAD2/3 Phosphorylation Motif Mutants in Cancer / TGFBR1 KD Mutants in Cancer / heart valve morphogenesis / positive regulation of vasculature development / hyaluronan catabolic process / regulation of transforming growth factor beta receptor signaling pathway / ATP biosynthetic process / receptor catabolic process / negative regulation of extracellular matrix disassembly / positive regulation of extracellular matrix assembly / type II transforming growth factor beta receptor binding / TGFBR1 LBD Mutants in Cancer / positive regulation of chemotaxis / negative regulation of biomineral tissue development / type I transforming growth factor beta receptor binding / cell-cell junction organization / negative regulation of myoblast differentiation / positive regulation of vascular permeability / deubiquitinase activator activity / response to cholesterol / positive regulation of endothelial cell apoptotic process / positive regulation of chemokine (C-X-C motif) ligand 2 production / aortic valve morphogenesis / positive regulation of fibroblast migration / phosphate-containing compound metabolic process / negative regulation of protein localization to plasma membrane / membrane protein intracellular domain proteolysis / SMAD2/3 Phosphorylation Motif Mutants in Cancer / TGFBR1 KD Mutants in Cancer / heart valve morphogenesis / positive regulation of vasculature development / hyaluronan catabolic process / regulation of transforming growth factor beta receptor signaling pathway / ATP biosynthetic process / receptor catabolic process / negative regulation of extracellular matrix disassembly / positive regulation of extracellular matrix assembly / type II transforming growth factor beta receptor binding / TGFBR1 LBD Mutants in Cancer / positive regulation of chemotaxis / negative regulation of biomineral tissue development / type I transforming growth factor beta receptor binding / cell-cell junction organization / negative regulation of myoblast differentiation / positive regulation of vascular permeability / deubiquitinase activator activity / response to cholesterol / positive regulation of endothelial cell apoptotic process / positive regulation of chemokine (C-X-C motif) ligand 2 production / aortic valve morphogenesis / positive regulation of fibroblast migration / phosphate-containing compound metabolic process / negative regulation of protein localization to plasma membrane /  sprouting angiogenesis / neural tube development / Molecules associated with elastic fibres / RUNX3 regulates CDKN1A transcription / positive regulation of epidermal growth factor receptor signaling pathway / ventricular cardiac muscle tissue morphogenesis / macrophage derived foam cell differentiation / negative regulation of fat cell differentiation / Syndecan interactions / negative regulation of cell-cell adhesion / positive regulation of interleukin-17 production / TGF-beta receptor signaling activates SMADs / negative regulation of cell differentiation / positive regulation of SMAD protein signal transduction / RUNX3 regulates p14-ARF / positive regulation of cell division / cellular response to low-density lipoprotein particle stimulus / negative regulation of blood vessel endothelial cell migration / negative regulation of cell cycle / ECM proteoglycans / positive regulation of vascular endothelial growth factor production / positive regulation of collagen biosynthetic process / sprouting angiogenesis / neural tube development / Molecules associated with elastic fibres / RUNX3 regulates CDKN1A transcription / positive regulation of epidermal growth factor receptor signaling pathway / ventricular cardiac muscle tissue morphogenesis / macrophage derived foam cell differentiation / negative regulation of fat cell differentiation / Syndecan interactions / negative regulation of cell-cell adhesion / positive regulation of interleukin-17 production / TGF-beta receptor signaling activates SMADs / negative regulation of cell differentiation / positive regulation of SMAD protein signal transduction / RUNX3 regulates p14-ARF / positive regulation of cell division / cellular response to low-density lipoprotein particle stimulus / negative regulation of blood vessel endothelial cell migration / negative regulation of cell cycle / ECM proteoglycans / positive regulation of vascular endothelial growth factor production / positive regulation of collagen biosynthetic process /  epithelial to mesenchymal transition / positive regulation of epithelial to mesenchymal transition / positive regulation of blood vessel endothelial cell migration / epithelial to mesenchymal transition / positive regulation of epithelial to mesenchymal transition / positive regulation of blood vessel endothelial cell migration /  vasculogenesis / lymph node development / chondrocyte differentiation / hematopoietic progenitor cell differentiation / salivary gland morphogenesis / extrinsic apoptotic signaling pathway / positive regulation of protein dephosphorylation / vasculogenesis / lymph node development / chondrocyte differentiation / hematopoietic progenitor cell differentiation / salivary gland morphogenesis / extrinsic apoptotic signaling pathway / positive regulation of protein dephosphorylation /  regulation of cell migration / cellular response to transforming growth factor beta stimulus / regulation of cell migration / cellular response to transforming growth factor beta stimulus /  antigen binding / positive regulation of protein metabolic process / protein export from nucleus / Downregulation of TGF-beta receptor signaling / antigen binding / positive regulation of protein metabolic process / protein export from nucleus / Downregulation of TGF-beta receptor signaling /  extracellular matrix / transforming growth factor beta receptor signaling pathway / positive regulation of superoxide anion generation / negative regulation of miRNA transcription / TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) / negative regulation of protein phosphorylation / platelet alpha granule lumen / response to progesterone / extracellular matrix / transforming growth factor beta receptor signaling pathway / positive regulation of superoxide anion generation / negative regulation of miRNA transcription / TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) / negative regulation of protein phosphorylation / platelet alpha granule lumen / response to progesterone /  cytokine activity / neural tube closure / positive regulation of protein secretion / positive regulation of protein-containing complex assembly cytokine activity / neural tube closure / positive regulation of protein secretion / positive regulation of protein-containing complex assemblySimilarity search - Function |
| Biological species |   Homo sapiens (human) Homo sapiens (human) |
 Citation Citation |  Date: 2019 Jul 1 Date: 2019 Jul 1Title: Structural consequences of transforming growth factor beta-1 activation from near-therapeutic X-ray doses Authors: Stachowski T / Grant T |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDFD2 SASDFD2 |
|---|
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
- Sample
Sample
 Sample Sample | Name: wild-type human Latent Transforming Growth Factor beta 1 (LTGFB-1) Specimen concentration: 1 mg/ml |
|---|---|
| Buffer | Name: phosphate buffered saline 2% glycerol / pH: 7.4 |
| Entity #1478 | Name: LTGFB-1 / Type: protein / Description: Human Latent Transforming Growth Factor beta 1 / Formula weight: 42.958 / Num. of mol.: 2 / Source: Homo sapiens / References: UniProt: P01137 Sequence: HHHHHHLEVL FQGPLSTSKT IDMELVKRKR IEAIRGQILS KLRLASPPSQ GEVPPGPLPE AVLALYNSTR DRVAGESAEP EPEPEADYYA KEVTRVLMVE THNEIYDKFK QSTHSIYMFF NTSELREAVP EPVLLSRAEL RLLRLKLKVE QHVELYQKYS NNSWRYLSNR ...Sequence: HHHHHHLEVL FQGPLSTSKT IDMELVKRKR IEAIRGQILS KLRLASPPSQ GEVPPGPLPE AVLALYNSTR DRVAGESAEP EPEPEADYYA KEVTRVLMVE THNEIYDKFK QSTHSIYMFF NTSELREAVP EPVLLSRAEL RLLRLKLKVE QHVELYQKYS NNSWRYLSNR LLAPSDSPEW LSFDVTGVVR QWLSRGGEIE GFRLSAHCSC DSRDNTLQVD INGFTTGRRG DLATIHGMNR PFLLLMATPL ERAQHLQSSR HRRALDTNYC FSSTEKNCCV RQLYIDFRKD LGWKWIHEPK GYHANFCLGP CPYIWSLDTQ YSKVLALYNQ HNPGASAAPC CVPQALEPLP IVYYVGRKPK VEQLSNMIVR SCKCS |
-Experimental information
| Beam | Instrument name: Advanced Light Source (ALS) 12.3.1 (SIBYLS) City: Berkeley, CA / 国: USA  / Type of source: X-ray synchrotron / Type of source: X-ray synchrotron Synchrotron / Wavelength: 0.1127 Å / Dist. spec. to detc.: 2 mm Synchrotron / Wavelength: 0.1127 Å / Dist. spec. to detc.: 2 mm | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus3 X 2M / Pixsize x: 172 mm | ||||||||||||||||||||||||||||||
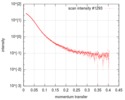
| Scan | Measurement date: Oct 4, 2018 / Storage temperature: 4 °C / Cell temperature: 10 °C / Exposure time: 0.1 sec. / Number of frames: 49 / Unit: 1/A /
| ||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| ||||||||||||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller