[English] 日本語
 Yorodumi
Yorodumi- SASDF44: RXR/RAR Heterodimer : N-CoRNID Complex (1:1) (Nuclear receptor ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 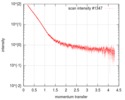 |
|---|---|
 Sample Sample | RXR/RAR Heterodimer : N-CoRNID Complex (1:1)
|
| Function / homology |  Function and homology information Function and homology informationTranscriptional regulation of granulopoiesis / Carnitine metabolism / Transcriptional regulation of white adipocyte differentiation / Regulation of pyruvate dehydrogenase (PDH) complex / Sertoli cell fate commitment / Signaling by Retinoic Acid / NR1H2 & NR1H3 regulate gene expression to control bile acid homeostasis / definitive erythrocyte differentiation / NR1H3 & NR1H2 regulate gene expression linked to cholesterol transport and efflux / positive regulation of binding ...Transcriptional regulation of granulopoiesis / Carnitine metabolism / Transcriptional regulation of white adipocyte differentiation / Regulation of pyruvate dehydrogenase (PDH) complex / Sertoli cell fate commitment / Signaling by Retinoic Acid / NR1H2 & NR1H3 regulate gene expression to control bile acid homeostasis / definitive erythrocyte differentiation / NR1H3 & NR1H2 regulate gene expression linked to cholesterol transport and efflux / positive regulation of binding / CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation / SUMOylation of intracellular receptors / trachea cartilage development / Downregulation of SMAD2/3:SMAD4 transcriptional activity / Recycling of bile acids and salts / Synthesis of bile acids and bile salts / Nuclear Receptor transcription pathway / ventricular cardiac muscle cell differentiation / visceral serous pericardium development / Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol / Synthesis of bile acids and bile salts via 27-hydroxycholesterol / HDACs deacetylate histones / mesenchyme development / chondroblast differentiation / positive regulation of translational initiation by iron / Endogenous sterols / embryonic camera-type eye development / Notch-HLH transcription pathway / glandular epithelial cell development / maternal placenta development / negative regulation of granulocyte differentiation /  protein kinase B binding / cellular response to corticotropin-releasing hormone stimulus / growth plate cartilage development / anatomical structure development / positive regulation of T-helper 2 cell differentiation / prostate gland development / secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development / negative regulation of cartilage development / Regulation of lipid metabolism by PPARalpha / thalamus development / retinoic acid-responsive element binding / regulation of hematopoietic progenitor cell differentiation / Cytoprotection by HMOX1 / positive regulation of thyroid hormone mediated signaling pathway / angiogenesis involved in coronary vascular morphogenesis / positive regulation of interleukin-5 production / nuclear glucocorticoid receptor binding / cardiac muscle cell differentiation / positive regulation of interleukin-13 production / nuclear retinoic acid receptor binding / protein kinase B binding / cellular response to corticotropin-releasing hormone stimulus / growth plate cartilage development / anatomical structure development / positive regulation of T-helper 2 cell differentiation / prostate gland development / secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development / negative regulation of cartilage development / Regulation of lipid metabolism by PPARalpha / thalamus development / retinoic acid-responsive element binding / regulation of hematopoietic progenitor cell differentiation / Cytoprotection by HMOX1 / positive regulation of thyroid hormone mediated signaling pathway / angiogenesis involved in coronary vascular morphogenesis / positive regulation of interleukin-5 production / nuclear glucocorticoid receptor binding / cardiac muscle cell differentiation / positive regulation of interleukin-13 production / nuclear retinoic acid receptor binding /  ion binding / camera-type eye development / ion binding / camera-type eye development /  retinoic acid binding / response to vitamin A / positive regulation of vitamin D receptor signaling pathway / regulation of thyroid hormone mediated signaling pathway / peroxisome proliferator activated receptor binding / nuclear vitamin D receptor binding / apoptotic cell clearance / retinoic acid binding / response to vitamin A / positive regulation of vitamin D receptor signaling pathway / regulation of thyroid hormone mediated signaling pathway / peroxisome proliferator activated receptor binding / nuclear vitamin D receptor binding / apoptotic cell clearance /  limb development / histone deacetylase regulator activity / nuclear thyroid hormone receptor binding / limb development / histone deacetylase regulator activity / nuclear thyroid hormone receptor binding /  protein kinase A binding / cardiac muscle cell proliferation / ureteric bud development / protein kinase A binding / cardiac muscle cell proliferation / ureteric bud development /  regulation of myelination / ventricular cardiac muscle tissue morphogenesis / Signaling by Retinoic Acid / DNA-binding transcription repressor activity / regulation of myelination / ventricular cardiac muscle tissue morphogenesis / Signaling by Retinoic Acid / DNA-binding transcription repressor activity /  DNA binding domain binding / nuclear steroid receptor activity / RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding / regulation of branching involved in prostate gland morphogenesis / DNA binding domain binding / nuclear steroid receptor activity / RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding / regulation of branching involved in prostate gland morphogenesis /  heterocyclic compound binding / LBD domain binding / positive regulation of interleukin-4 production / face development / alpha-actinin binding / germ cell development / locomotor rhythm / negative regulation of type II interferon production / negative regulation of tumor necrosis factor production / cellular response to Thyroglobulin triiodothyronine / regulation of multicellular organism growth / epidermis development / retinoic acid receptor signaling pathway / negative regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / establishment of skin barrier / cellular response to estrogen stimulus / positive regulation of bone mineralization / positive regulation of cell cycle / nuclear retinoid X receptor binding / heart morphogenesis / cellular response to retinoic acid / response to retinoic acid / response to glucocorticoid / peroxisome proliferator activated receptor signaling pathway / transcription repressor complex / heterocyclic compound binding / LBD domain binding / positive regulation of interleukin-4 production / face development / alpha-actinin binding / germ cell development / locomotor rhythm / negative regulation of type II interferon production / negative regulation of tumor necrosis factor production / cellular response to Thyroglobulin triiodothyronine / regulation of multicellular organism growth / epidermis development / retinoic acid receptor signaling pathway / negative regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / establishment of skin barrier / cellular response to estrogen stimulus / positive regulation of bone mineralization / positive regulation of cell cycle / nuclear retinoid X receptor binding / heart morphogenesis / cellular response to retinoic acid / response to retinoic acid / response to glucocorticoid / peroxisome proliferator activated receptor signaling pathway / transcription repressor complex /  embryo implantation embryo implantationSimilarity search - Function |
| Biological species | Mouse (Mus musculus) Human (Homo sapiens) |
 Citation Citation |  Journal: Structure / Year: 2019 Journal: Structure / Year: 2019Title: Interplay of Protein Disorder in Retinoic Acid Receptor Heterodimer and Its Corepressor Regulates Gene Expression. Authors: Tiago N Cordeiro / Nathalie Sibille / Pierre Germain / Philippe Barthe / Abdelhay Boulahtouf / Fréderic Allemand / Rémy Bailly / Valérie Vivat / Christine Ebel / Alessandro Barducci / ...Authors: Tiago N Cordeiro / Nathalie Sibille / Pierre Germain / Philippe Barthe / Abdelhay Boulahtouf / Fréderic Allemand / Rémy Bailly / Valérie Vivat / Christine Ebel / Alessandro Barducci / William Bourguet / Albane le Maire / Pau Bernadó /    Abstract: In its unliganded form, the retinoic acid receptor (RAR) in heterodimer with the retinoid X receptor (RXR) exerts a strong repressive activity facilitated by the recruitment of transcriptional ...In its unliganded form, the retinoic acid receptor (RAR) in heterodimer with the retinoid X receptor (RXR) exerts a strong repressive activity facilitated by the recruitment of transcriptional corepressors in the promoter region of target genes. By integrating complementary structural, biophysical, and computational information, we demonstrate that intrinsic disorder is a required feature for the precise regulation of RAR activity. We show that structural dynamics of RAR and RXR H12 regions is an essential mechanism for RAR regulation. Unexpectedly we found that, while mainly disordered, the corepressor N-CoR presents evolutionary conserved structured regions involved in transient intramolecular contacts. In the presence of RXR/RAR, N-CoR exploits its multivalency to form a cooperative multisite complex that displays equilibrium between different conformational states that can be tuned by cognate ligands and receptor mutations. This equilibrium is key to preserving the repressive basal state while allowing the conversion to a transcriptionally active form. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDF44 SASDF44 |
|---|
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
- Sample
Sample
 Sample Sample | Name: RXR/RAR Heterodimer : N-CoRNID Complex (1:1) / Specimen concentration: 0.90-5.00 / Entity id: 1515 / 1536 / 1537 |
|---|---|
| Buffer | Name: 50mM Tris HCl, 150mM NaCl, 2mM TCEP. / pH: 7.5 |
| Entity #1515 | Name: N-CoR-NID / Type: protein Description: Nuclear receptor CoRepressor 1; Nuclear Receptor Interaction Domain (NID) Formula weight: 29.139 / Num. of mol.: 1 / Source: Mouse (Mus musculus) / References: UniProt: Q60974 Sequence: GPHMQVPRTH RLITLADHIC QIITQDFARN QVPSQASTST FQTSPSALSS TPVRTKTSSR YSPESQSQTV LHPRPGPRVS PENLVDKSRG SRPGKSPERS HIPSEPYEPI SPPQGPAVHE KQDSMLLLSQ RGVDPAEQRS DSRSPGSISY LPSFFTKLES TSPMVKSKKQ ...Sequence: GPHMQVPRTH RLITLADHIC QIITQDFARN QVPSQASTST FQTSPSALSS TPVRTKTSSR YSPESQSQTV LHPRPGPRVS PENLVDKSRG SRPGKSPERS HIPSEPYEPI SPPQGPAVHE KQDSMLLLSQ RGVDPAEQRS DSRSPGSISY LPSFFTKLES TSPMVKSKKQ EIFRKLNSSG GGDSDMAAAQ PGTEIFNLPA VTTSGAVSSR SHSFADPASN LGLEDIIRKA LMGSFDDKVE DHGVVMSHPV GIMPGSASTS VVTSSEARRD E |
| Entity #1536 | Name: RXR / Type: protein Description: Retinoid-X receptor alpha (RXR-alpha) Ligand Binding Domain (LBD) Formula weight: 26.47 / Num. of mol.: 1 / Source: Mouse (Mus musculus) / References: UniProt: P28700 Sequence: SANEDMPVEK ILEAELAVEP KTETYVEANM GLNPSSPNDP VTNICQAADK QLFTLVEWAK RIPHFSELPL DDQVILLRAG WNELLIASAS HRSIAVKDGI LLATGLHVHR NSAHSAGVGA IFDRVLTELV SKMRDMQMDK TELGCLRAIV LFNPDSKGLS NPAEVEALRE ...Sequence: SANEDMPVEK ILEAELAVEP KTETYVEANM GLNPSSPNDP VTNICQAADK QLFTLVEWAK RIPHFSELPL DDQVILLRAG WNELLIASAS HRSIAVKDGI LLATGLHVHR NSAHSAGVGA IFDRVLTELV SKMRDMQMDK TELGCLRAIV LFNPDSKGLS NPAEVEALRE KVYASLEAYC KHKYPEQPGR FAKLLLRLPA LRSIGLKCLE HLFFFKLIGD TPIDTFLMEM LEAPHQAT |
| Entity #1537 | Name: RAR / Type: protein Description: Retinoic acid receptor alpha (RAR-alpha) Ligand binding domain (LDB) Formula weight: 28.047 / Num. of mol.: 1 / Source: Human (Homo sapiens) / References: UniProt: P10276 Sequence: GSHESYTLTP EVGELIEKVR KAHQETFPAL CQLGKYTTNN SSEQRVSLDI DLWDKFSELS TKCIIKTVEF AKQLPGFTTL TIADQITLLK AACLDILILR ICTRYTPEQD TMTFSDGLTL NRTQMHNAGF GPLTDLVFAF ANQLLPLEMD DAETGLLSAI CLICGDRQDL ...Sequence: GSHESYTLTP EVGELIEKVR KAHQETFPAL CQLGKYTTNN SSEQRVSLDI DLWDKFSELS TKCIIKTVEF AKQLPGFTTL TIADQITLLK AACLDILILR ICTRYTPEQD TMTFSDGLTL NRTQMHNAGF GPLTDLVFAF ANQLLPLEMD DAETGLLSAI CLICGDRQDL EQPDRVDMLQ EPLLEALKVY VRKRRPSRPH MFPKMLMKIT DLRSISAKGA ERVITLKMEI PGSMPPLIQE MLENSEGLD |
-Experimental information
| Beam | Instrument name: ESRF BM29 / City: Grenoble / 国: France  / Type of source: X-ray synchrotron / Type of source: X-ray synchrotron Synchrotron / Wavelength: 0.099 Å / Dist. spec. to detc.: 2.9 mm Synchrotron / Wavelength: 0.099 Å / Dist. spec. to detc.: 2.9 mm | |||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 1M / Type: Dectris / Pixsize x: 172 mm | |||||||||||||||||||||||||||||||||||||||
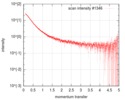
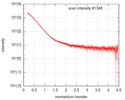
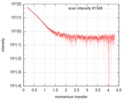
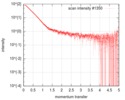
| Scan |
| |||||||||||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| |||||||||||||||||||||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller