[English] 日本語
 Yorodumi
Yorodumi- SASDD25: 1:1 Mixture between Protein sex-lethal mutant (Sxl10GS) and RNA d... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDD25 |
|---|---|
 Sample Sample | 1:1 Mixture between Protein sex-lethal mutant (Sxl10GS) and RNA decaneucleotide UGU8
|
| Function / homology |  Function and homology information Function and homology informationsex determination, primary response to X:A ratio / germarium-derived cystoblast division / epithelium regeneration / female sex determination / somatic sex determination / female germ-line sex determination / oocyte differentiation / imaginal disc growth /  sex-chromosome dosage compensation / regulation of stem cell division ...sex determination, primary response to X:A ratio / germarium-derived cystoblast division / epithelium regeneration / female sex determination / somatic sex determination / female germ-line sex determination / oocyte differentiation / imaginal disc growth / sex-chromosome dosage compensation / regulation of stem cell division ...sex determination, primary response to X:A ratio / germarium-derived cystoblast division / epithelium regeneration / female sex determination / somatic sex determination / female germ-line sex determination / oocyte differentiation / imaginal disc growth /  sex-chromosome dosage compensation / regulation of stem cell division / sex determination / poly-pyrimidine tract binding / sex-chromosome dosage compensation / regulation of stem cell division / sex determination / poly-pyrimidine tract binding /  messenger ribonucleoprotein complex / messenger ribonucleoprotein complex /  sex differentiation / alternative mRNA splicing, via spliceosome / negative regulation of receptor signaling pathway via JAK-STAT / sex differentiation / alternative mRNA splicing, via spliceosome / negative regulation of receptor signaling pathway via JAK-STAT /  poly(A) binding / poly(A) binding /  pre-mRNA binding / positive regulation of smoothened signaling pathway / reciprocal meiotic recombination / pre-mRNA binding / positive regulation of smoothened signaling pathway / reciprocal meiotic recombination /  regulation of mRNA splicing, via spliceosome / poly(U) RNA binding / regulation of mRNA splicing, via spliceosome / poly(U) RNA binding /  oogenesis / regulation of alternative mRNA splicing, via spliceosome / negative regulation of mRNA splicing, via spliceosome / negative regulation of translational initiation / mRNA regulatory element binding translation repressor activity / positive regulation of RNA splicing / mRNA 3'-UTR binding / mRNA 5'-UTR binding / negative regulation of translation / protein stabilization / molecular adaptor activity / oogenesis / regulation of alternative mRNA splicing, via spliceosome / negative regulation of mRNA splicing, via spliceosome / negative regulation of translational initiation / mRNA regulatory element binding translation repressor activity / positive regulation of RNA splicing / mRNA 3'-UTR binding / mRNA 5'-UTR binding / negative regulation of translation / protein stabilization / molecular adaptor activity /  ribonucleoprotein complex / protein-containing complex / ribonucleoprotein complex / protein-containing complex /  RNA binding / RNA binding /  nucleus / nucleus /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function |
| Biological species |   Drosophila melanogaster (fruit fly) Drosophila melanogaster (fruit fly)synthetic construct (others) |
 Citation Citation |  Journal: ACS Comb Sci / Year: 2018 Journal: ACS Comb Sci / Year: 2018Title: A General Small-Angle X-ray Scattering-Based Screening Protocol Validated for Protein-RNA Interactions. Authors: Po-Chia Chen / Pawel Masiewicz / Vladimir Rybin / Dmitri Svergun / Janosch Hennig /  Abstract: We present a screening protocol utilizing small-angle X-ray scattering (SAXS) to obtain structural information on biomolecular interactions independent of prior knowledge, so as to complement ...We present a screening protocol utilizing small-angle X-ray scattering (SAXS) to obtain structural information on biomolecular interactions independent of prior knowledge, so as to complement affinity-based screening and provide leads for further exploration. This protocol categorizes ligand titrations by computing pairwise agreement between curves, and separately estimates affinities by quantifying complex formation as a departure from the linear sum properties of solution SAXS. The protocol is validated by sparse sequence search around the native poly uridine RNA motifs of the two-RRM domain Sex-lethal protein (Sxl). The screening of 35 RNA motifs between 4 to 10 nucleotides reveals a strong variation of resulting complexes, revealed to be preference-switching between 1:1 and 2:2 binding stoichiometries upon addition of structural modeling. Validation of select sequences in isothermal calorimetry and NMR titration retrieves domain-specific roles and function of a guanine anchor. These findings reinforce the suitability of SAXS as a complement in lead identification. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Models
| Model #1877 |  Type: mix / Software: (2.1) / Radius of dummy atoms: 1.90 A / Chi-square value: 0.777 / P-value: 0.240677  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|---|
| Model #1878 |  Type: mix / Software: (2.1) / Radius of dummy atoms: 1.90 A / Chi-square value: 0.777 / P-value: 0.240677  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
| Model #1879 |  Type: mix / Software: (2.1) / Radius of dummy atoms: 1.90 A / Chi-square value: 0.777 / P-value: 0.240677  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
- Sample
Sample
 Sample Sample | Name: 1:1 Mixture between Protein sex-lethal mutant (Sxl10GS) and RNA decaneucleotide UGU8 Specimen concentration: 1 mg/ml / Entity id: 1008 / 1009 |
|---|---|
| Buffer | Name: 50% dilution of protein buffer {10 mM KP, 50 mM NaCl, 10 mM DTT pH 6} with {milliQ-water pH 7} suspended RNA pH: 6 |
| Entity #1008 | Name: Sxl10GS / Type: protein / Description: Protein sex-lethal mutant / Formula weight: 20.432 / Num. of mol.: 2 / Source: Drosophila melanogaster / References: UniProt: P19339 Sequence: GAMASNTNLI VNYLPQDMTD RELYALFRAI GPINTCRIMR DYKTGYSFGY AFVDFTSEMD SQRAIKVLNG ITVRNKRLKV SYARPGGGSG SGGGGSGESI KDTNLYVTNL PRTITDDQLD TIFGKYGSIV QKNILRDKLT GRPRGVAFVR YNKREEAQEA ISALNNVIPE GGSQPLSVRL AEEHGK |
| Entity #1009 | Name: UGU8 / Type: RNA / Description: RNA decaneucleotide UGU8 / Formula weight: 3.119 / Num. of mol.: 2 / Source: synthetic construct Sequence: UGUUUUUUUU |
-Experimental information
| Beam | Instrument name: ESRF BM29 / City: Grenoble / 国: France  / Type of source: X-ray synchrotron / Type of source: X-ray synchrotron Synchrotron / Wavelength: 0.09919 Å / Dist. spec. to detc.: 2.867 mm Synchrotron / Wavelength: 0.09919 Å / Dist. spec. to detc.: 2.867 mm | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 1M | |||||||||||||||||||||||||||||||||
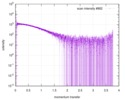
| Scan | Measurement date: Apr 12, 2017 / Storage temperature: 20 °C / Cell temperature: 20 °C / Exposure time: 0.5 sec. / Number of frames: 10 / Unit: 1/nm /
| |||||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| |||||||||||||||||||||||||||||||||
| Result | Comments: This synchrotron session suffered from misalignment of the beamstop resulting in parasitic scattering at low angles. Note that the uploaded EOM ensemble is chosen as the 2:2-complex with ...Comments: This synchrotron session suffered from misalignment of the beamstop resulting in parasitic scattering at low angles. Note that the uploaded EOM ensemble is chosen as the 2:2-complex with one RNA monomer as fully bound by two Sxl10GS molecules in the canonical position from Protein data bankk (PDB) entry 1B7F.pdb. The second RNA monomer is placed in the canonical position bound to the free RRM2, resulting in three rigid bodies: RRM2+RNA, RRM1+RRM2+RNA, and RRM1.
|
 Movie
Movie Controller
Controller

 SASDD25
SASDD25