[English] 日本語
 Yorodumi
Yorodumi- SASDBL2: MBP-PICK1 fusion (Maltose Binding Protein fused to Protein Intera... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDBL2 |
|---|---|
 Sample Sample | MBP-PICK1 fusion
|
| Function / homology |  Function and homology information Function and homology informationmembrane curvature sensor activity / postsynaptic early endosome / glial cell development / neuronal ion channel clustering / cellular response to decreased oxygen levels /  Arp2/3 complex binding / Trafficking of GluR2-containing AMPA receptors / epigenetic programming of gene expression / regulation of Arp2/3 complex-mediated actin nucleation / negative regulation of Arp2/3 complex-mediated actin nucleation ...membrane curvature sensor activity / postsynaptic early endosome / glial cell development / neuronal ion channel clustering / cellular response to decreased oxygen levels / Arp2/3 complex binding / Trafficking of GluR2-containing AMPA receptors / epigenetic programming of gene expression / regulation of Arp2/3 complex-mediated actin nucleation / negative regulation of Arp2/3 complex-mediated actin nucleation ...membrane curvature sensor activity / postsynaptic early endosome / glial cell development / neuronal ion channel clustering / cellular response to decreased oxygen levels /  Arp2/3 complex binding / Trafficking of GluR2-containing AMPA receptors / epigenetic programming of gene expression / regulation of Arp2/3 complex-mediated actin nucleation / negative regulation of Arp2/3 complex-mediated actin nucleation / monoamine transport / Arp2/3 complex binding / Trafficking of GluR2-containing AMPA receptors / epigenetic programming of gene expression / regulation of Arp2/3 complex-mediated actin nucleation / negative regulation of Arp2/3 complex-mediated actin nucleation / monoamine transport /  genomic imprinting / protein kinase C-activating G protein-coupled receptor signaling pathway / retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum / dendritic spine organization / long-term synaptic depression / genomic imprinting / protein kinase C-activating G protein-coupled receptor signaling pathway / retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum / dendritic spine organization / long-term synaptic depression /  dendritic spine maintenance / receptor clustering / regulation of insulin secretion / positive regulation of receptor internalization / cellular response to glucose starvation / trans-Golgi network membrane / dendritic spine maintenance / receptor clustering / regulation of insulin secretion / positive regulation of receptor internalization / cellular response to glucose starvation / trans-Golgi network membrane /  protein kinase C binding / G protein-coupled receptor binding / Cell surface interactions at the vascular wall / protein kinase C binding / G protein-coupled receptor binding / Cell surface interactions at the vascular wall /  intracellular protein transport / intracellular protein transport /  phospholipid binding / endocytic vesicle membrane / phospholipid binding / endocytic vesicle membrane /  actin filament binding / actin filament binding /  synaptic vesicle / synaptic vesicle /  presynaptic membrane / presynaptic membrane /  postsynaptic density / postsynaptic density /  cytoskeleton / neuron projection / protein domain specific binding / cytoskeleton / neuron projection / protein domain specific binding /  protein phosphorylation / protein phosphorylation /  signaling receptor binding / signaling receptor binding /  synapse / perinuclear region of cytoplasm / synapse / perinuclear region of cytoplasm /  Golgi apparatus / identical protein binding / Golgi apparatus / identical protein binding /  metal ion binding / metal ion binding /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function |
| Biological species |   Homo sapiens (human) Homo sapiens (human) |
 Citation Citation |  Journal: Mol Biol Cell / Year: 2015 Journal: Mol Biol Cell / Year: 2015Title: PICK1 is implicated in organelle motility in an Arp2/3 complex-independent manner. Authors: Yadaiah Madasu / Changsong Yang / Malgorzata Boczkowska / Kelley A Bethoney / Adam Zwolak / Grzegorz Rebowski / Tatyana Svitkina / Roberto Dominguez /  Abstract: PICK1 is a modular scaffold implicated in synaptic receptor trafficking. It features a PDZ domain, a BAR domain, and an acidic C-terminal tail (ACT). Analysis by small- angle x-ray scattering ...PICK1 is a modular scaffold implicated in synaptic receptor trafficking. It features a PDZ domain, a BAR domain, and an acidic C-terminal tail (ACT). Analysis by small- angle x-ray scattering suggests a structural model that places the receptor-binding site of the PDZ domain and membrane-binding surfaces of the BAR and PDZ domains adjacent to each other on the concave side of the banana-shaped PICK1 dimer. In the model, the ACT of one subunit of the dimer interacts with the PDZ and BAR domains of the other subunit, possibly accounting for autoinhibition. Consistently, full-length PICK1 shows diffuse cytoplasmic localization, but it clusters on vesicle-like structures that colocalize with the trans-Golgi network marker TGN38 upon deletion of either the ACT or PDZ domain. This localization is driven by the BAR domain. Live-cell imaging further reveals that PICK1-associated vesicles undergo fast, nondirectional motility in an F-actin-dependent manner, but deleting the ACT dramatically reduces vesicle speed. Thus the ACT links PICK1-associated vesicles to a motility factor, likely myosin, but, contrary to previous reports, PICK1 neither binds nor inhibits Arp2/3 complex. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDBL2 SASDBL2 |
|---|
-Related structure data
| Similar structure data |
|---|
- External links
External links
| Related items in Molecule of the Month |
|---|
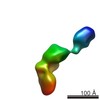
-Models
| Model #402 |  Type: dummy / Software: crysol / Radius of dummy atoms: 1.90 A / Chi-square value: 1.1881  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|
- Sample
Sample
 Sample Sample | Name: MBP-PICK1 fusion / Specimen concentration: 0.46-7.50 |
|---|---|
| Buffer | Name: 50 mM Tris / pH: 7.5 / Composition: 300 mM NaCl, 1 mM maltose, 1 mM EGTA, 2 mM DTT |
| Entity #266 | Name: MBP-PICK1 / Type: protein Description: Maltose Binding Protein fused to Protein Interacting with C kinase 1 Formula weight: 87.089 / Num. of mol.: 2 / Source: Homo sapiens / References: UniProt: Q9NRD5 Sequence: MKIEEGKLVI WINGDKGYNG LAEVGKKFEK DTGIKVTVEH PDKLEEKFPQ VAATGDGPDI IFWAHDRFGG YAQSGLLAEI TPDKAFQDKL YPFTWDAVRY NGKLIAYPIA VEALSLIYNK DLLPNPPKTW EEIPALDKEL KAKGKSALMF NLQEPYFTWP LIAADGGYAF ...Sequence: MKIEEGKLVI WINGDKGYNG LAEVGKKFEK DTGIKVTVEH PDKLEEKFPQ VAATGDGPDI IFWAHDRFGG YAQSGLLAEI TPDKAFQDKL YPFTWDAVRY NGKLIAYPIA VEALSLIYNK DLLPNPPKTW EEIPALDKEL KAKGKSALMF NLQEPYFTWP LIAADGGYAF KYENGKYDIK DVGVDNAGAK AGLTFLVDLI KNKHMNADTD YSIAEAAFNK GETAMTINGP WAWSNIDTSK VNYGVTVLPT FKGQPSKPFV GVLSAGINAA SPNKELAKEF LENYLLTDEG LEAVNKDKPL GAVALKSYEE ELAKDPRIAA TMENAQKGEI MPNIPQMSAF WYAVRTAVIN AASGRQTVDA ALAAAQTNAA AMFADLDYDI EEDKLGIPTV PGKVTLQKDA QNLIGISIGG GAQYCPCLYI VQVFDNTPAA LDGTVAAGDE ITGVNGRSIK GKTKVEVAKM IQEVKGEVTI HYNKLQADPK QGMSLDIVLK KVKHRLVENM SSGTADALGL SRAILCNDGL VKRLEELERT AELYKGMTEH TKNLLRAFYE LSQTHRAFGD VFSVIGVREP QPAASEAFVK FADAHRSIEK FGIRLLKTIK PMLTDLNTYL NKAIPDTRLT IKKYLDVKFE YLSYCLKVKE MDDEEYSCIA LGEPLYRVST GNYEYRLILR CRQEARARFS QMRKDVLEKM ELLDQKHVQD IVFQLQRLVS TMSKYYNDCY AVLRDADVFP IEVDLAHTTL AYGLNQEEFT DGEEEEEEED TAAGEPSRDT RGAAGPLDKG GSWCDS |
-Experimental information
| Beam | Instrument name: Cornell High Energy Synchrotron Source (CHESS) G1 City: Ithaca, NY / 国: USA  / Type of source: X-ray synchrotron / Type of source: X-ray synchrotron Synchrotron Synchrotron | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Finger Lakes CCD / Type: CCD | ||||||||||||||||||
| Scan |
| ||||||||||||||||||
| Distance distribution function P(R) |
| ||||||||||||||||||
| Result | Comments: This dataset was collected at a concentration of 3.75 mg/ml.
|
 Movie
Movie Controller
Controller