[English] 日本語
 Yorodumi
Yorodumi- SASDB28: Complement factor 1q (C1q) (Complement C1q subcomponent subunit C... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 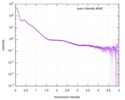 |
|---|---|
 Sample Sample | Complement factor 1q (C1q)
|
| Function / homology |  Function and homology information Function and homology informationcomplement component C1 complex / complement component C1q complex / negative regulation of macrophage differentiation / synapse pruning / negative regulation of granulocyte differentiation / vertebrate eye-specific patterning / complement-mediated synapse pruning / collagen trimer /  complement activation / Classical antibody-mediated complement activation ...complement component C1 complex / complement component C1q complex / negative regulation of macrophage differentiation / synapse pruning / negative regulation of granulocyte differentiation / vertebrate eye-specific patterning / complement-mediated synapse pruning / collagen trimer / complement activation / Classical antibody-mediated complement activation ...complement component C1 complex / complement component C1q complex / negative regulation of macrophage differentiation / synapse pruning / negative regulation of granulocyte differentiation / vertebrate eye-specific patterning / complement-mediated synapse pruning / collagen trimer /  complement activation / Classical antibody-mediated complement activation / neuron remodeling / Initial triggering of complement / complement activation / Classical antibody-mediated complement activation / neuron remodeling / Initial triggering of complement /  complement activation, classical pathway / complement activation, classical pathway /  Regulation of Complement cascade / astrocyte activation / synapse organization / microglial cell activation / cell-cell signaling / Regulation of Complement cascade / astrocyte activation / synapse organization / microglial cell activation / cell-cell signaling /  amyloid-beta binding / postsynapse / collagen-containing extracellular matrix / blood microparticle / amyloid-beta binding / postsynapse / collagen-containing extracellular matrix / blood microparticle /  immune response / immune response /  innate immune response / innate immune response /  synapse / synapse /  extracellular space / extracellular region extracellular space / extracellular regionSimilarity search - Function |
| Biological species | Homo sapiens (plasma purified)  Homo sapiens (human) Homo sapiens (human) |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDB28 SASDB28 |
|---|
-Related structure data
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
|---|
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
- Sample
Sample
 Sample Sample | Name: Complement factor 1q (C1q) / Specimen concentration: 3.66-3.66 / Entity id: 541 / 542 / 543 |
|---|---|
| Buffer | Name: 50 mM TrisHCl, 145 mM NaCl, 3 mM CaCl2 / pH: 7.4 |
| Entity #541 | Name: C1qc / Type: protein / Description: Complement C1q subcomponent subunit C / Formula weight: 23.687 / Num. of mol.: 6 / Source: Homo sapiens (plasma purified) / References: UniProt: P02747 Sequence: EDLCRAPDGK KGEAGRPGRR GRPGLKGEQG EPGAPGIRTG IQGLKGDQGE PGPSGNPGKV GYPGPSGPLG ARGIPGIKGT KGSPGNIKDQ PRPAFSAIRR NPPMGGNVVI FDTVITNQEE PYQNHSGRFV CTVPGYYYFT FQVLSQWEIC LSIVSSSRGQ VRRSLGFCDT ...Sequence: EDLCRAPDGK KGEAGRPGRR GRPGLKGEQG EPGAPGIRTG IQGLKGDQGE PGPSGNPGKV GYPGPSGPLG ARGIPGIKGT KGSPGNIKDQ PRPAFSAIRR NPPMGGNVVI FDTVITNQEE PYQNHSGRFV CTVPGYYYFT FQVLSQWEIC LSIVSSSRGQ VRRSLGFCDT TNKGLFQVVS GGMVLQLQQG DQVWVEKDPK KGHIYQGSEA DSVFSGFLIF PSA |
| Entity #542 | Name: C1qb / Type: protein / Description: Complement C1q subcomponent subunit B / Formula weight: 23.742 / Num. of mol.: 6 / Source: Homo sapiens (plasma purified) / References: UniProt: P02746 Sequence: QLSCTGPPAI PGIPGIPGTP GPDGQPGTPG IKGEKGLPGL AGDHGEFGEK GDPGIPGNPG KVGPKGPMGP KGGPGAPGAP GPKGESGDYK ATQKIAFSAT RTINVPLRRD QTIRFDHVIT NMNNNYEPRS GKFTCKVPGL YYFTYHASSR GNLCVNLMRG RERAQKVVTF ...Sequence: QLSCTGPPAI PGIPGIPGTP GPDGQPGTPG IKGEKGLPGL AGDHGEFGEK GDPGIPGNPG KVGPKGPMGP KGGPGAPGAP GPKGESGDYK ATQKIAFSAT RTINVPLRRD QTIRFDHVIT NMNNNYEPRS GKFTCKVPGL YYFTYHASSR GNLCVNLMRG RERAQKVVTF CDYAYNTFQV TTGGMVLKLE QGENVFLQAT DKNSLLGMEG ANSIFSGFLL FPDMEA |
| Entity #543 | Name: C1qa / Type: protein / Description: Complement C1q subcomponent subunit A / Formula weight: 23.687 / Num. of mol.: 6 / Source: Homo sapiens / References: UniProt: P02745 Sequence: EDLCRAPDGK KGEAGRPGRR GRPGLKGEQG EPGAPGIRTG IQGLKGDQGE PGPSGNPGKV GYPGPSGPLG ARGIPGIKGT KGSPGNIKDQ PRPAFSAIRR NPPMGGNVVI FDTVITNQEE PYQNHSGRFV CTVPGYYYFT FQVLSQWEIC LSIVSSSRGQ VRRSLGFCDT ...Sequence: EDLCRAPDGK KGEAGRPGRR GRPGLKGEQG EPGAPGIRTG IQGLKGDQGE PGPSGNPGKV GYPGPSGPLG ARGIPGIKGT KGSPGNIKDQ PRPAFSAIRR NPPMGGNVVI FDTVITNQEE PYQNHSGRFV CTVPGYYYFT FQVLSQWEIC LSIVSSSRGQ VRRSLGFCDT TNKGLFQVVS GGMVLQLQQG DQVWVEKDPK KGHIYQGSEA DSVFSGFLIF PSA |
-Experimental information
| Beam | Instrument name: ESRF BM29 / City: Grenoble / 国: France  / Type of source: X-ray synchrotron / Type of source: X-ray synchrotron Synchrotron / Wavelength: 0.0992 Å Synchrotron / Wavelength: 0.0992 Å | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 1M | |||||||||||||||||||||
| Scan |
| |||||||||||||||||||||
| Result | Type of curve: single_conc /
|
 Movie
Movie Controller
Controller




