+ Open data
Open data
- Basic information
Basic information
| Entry | 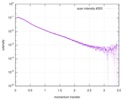 |
|---|---|
 Sample Sample | Cardiac myosin binding protein-C: domains C5-C6-C7
|
| Function / homology |  Function and homology information Function and homology informationC zone / regulation of muscle filament sliding / striated muscle myosin thick filament / cardiac myofibril / regulation of striated muscle contraction / positive regulation of ATP-dependent activity / Striated Muscle Contraction / A band / structural constituent of muscle / ventricular cardiac muscle tissue morphogenesis ...C zone / regulation of muscle filament sliding / striated muscle myosin thick filament / cardiac myofibril / regulation of striated muscle contraction / positive regulation of ATP-dependent activity / Striated Muscle Contraction / A band / structural constituent of muscle / ventricular cardiac muscle tissue morphogenesis / sarcomere organization /  myosin binding / myosin binding /  myosin heavy chain binding / ATPase activator activity / heart morphogenesis / cardiac muscle contraction / myosin heavy chain binding / ATPase activator activity / heart morphogenesis / cardiac muscle contraction /  titin binding / titin binding /  sarcomere / sarcomere /  actin binding / actin binding /  cell adhesion / identical protein binding / cell adhesion / identical protein binding /  metal ion binding / metal ion binding /  cytosol cytosolSimilarity search - Function |
| Biological species |   Homo sapiens (human) Homo sapiens (human) |
 Citation Citation |  Journal: Structure / Year: 2016 Journal: Structure / Year: 2016Title: Clinically Linked Mutations in the Central Domains of Cardiac Myosin-Binding Protein C with Distinct Phenotypes Show Differential Structural Effects. Authors: Naveed Ahmed Nadvi / Katharine A Michie / Ann H Kwan / J Mitchell Guss / Jill Trewhella /  Abstract: The structural effects of three missense mutations clinically linked to hypertrophic cardiomyopathy (HCM) and located in the central domains of cardiac myosin-binding protein C (cMyBP-C) have been ...The structural effects of three missense mutations clinically linked to hypertrophic cardiomyopathy (HCM) and located in the central domains of cardiac myosin-binding protein C (cMyBP-C) have been determined using small-angle scattering, infrared spectroscopy, and nuclear magnetic resonance spectroscopy. Bioinformatics and modeling were used to initially predict the expected structural impacts and assess the broader implications for function based on sequence conservation patterns. The experimental results generally affirm the predictions that two of the mutations (D745G, P873H) disrupt domain folding, while the third (R820Q) is likely to be entirely solvent exposed and thus more likely to have its impact through its interactions within the sarcomere. Each of the mutations is associated with distinct disease phenotypes, with respect to severity, stage of onset, and end phase. The results are discussed in terms of understanding key structural features of these domains essential for healthy function and the role they may play in disease development. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDB25 SASDB25 |
|---|
-Related structure data
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
|---|
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
- Sample
Sample
 Sample Sample | Name: Cardiac myosin binding protein-C: domains C5-C6-C7 / Specimen concentration: 0.70-3.40 |
|---|---|
| Buffer | Name: 25 mM Tris-HCl, 250 mM NaCl, 2 mM TCEP, 0.02% sodium azide Concentration: 25.00 mM / pH: 7.5 / Composition: 250 mM NaCl, 2 mM TCEP, 0.02% sodium azide |
| Entity #352 | Name: cMyBP-C / Type: protein Description: Cardiac myosin binding protein-C: domains C5-C6-C7 Formula weight: 35.751 / Num. of mol.: 1 / Source: Homo sapiens / References: UniProt: Q14896 Sequence: GPGSRQEPPK IHLDCPGRIP DTIVVVAGNK LRLDVPISGD PAPTVIWQKA ITQGNKAPAR PAPDAPEDTG DSDEWVFDKK LLCETEGRVR VETTKDRSIF TVEGAEKEDE GVYTVTVKNP VGEDQVNLTV KVIDVPDAPA APKISNVGED SCTVQWEPPA YDGGQPILGY ...Sequence: GPGSRQEPPK IHLDCPGRIP DTIVVVAGNK LRLDVPISGD PAPTVIWQKA ITQGNKAPAR PAPDAPEDTG DSDEWVFDKK LLCETEGRVR VETTKDRSIF TVEGAEKEDE GVYTVTVKNP VGEDQVNLTV KVIDVPDAPA APKISNVGED SCTVQWEPPA YDGGQPILGY ILERKKKKSY RWMRLNFDLI QELSHEARRM IEGVVYEMRV YAVNAIGMSR PSPASQPFMP IGPPSEPTHL AVEDVSDTTV SLKWRPPERV GAGGLDGYSV EYCPEGCSEW VAALQGLTEH TSILVKDLPT GARLLFRVRA HNMAGPGAPV TTTEPVTV |
-Experimental information
| Beam | Instrument name: Australian Synchrotron SAXS/WAXS / City: Melbourne / 国: Australia  / Shape / Shape : Point / Type of source: X-ray synchrotron : Point / Type of source: X-ray synchrotron Synchrotron / Dist. spec. to detc.: 1.6 mm Synchrotron / Dist. spec. to detc.: 1.6 mm | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 1M | |||||||||||||||||||||||||||||||||
| Scan |
| |||||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| |||||||||||||||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller







