[English] 日本語
 Yorodumi
Yorodumi- PDB-8t9x: Zophobas morio black wasting virus strain NJ2-molitor virion structure -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8t9x | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
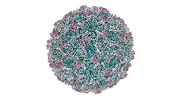
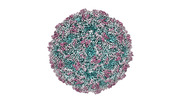
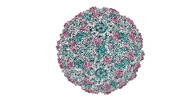
| Title | Zophobas morio black wasting virus strain NJ2-molitor virion structure | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | VIRUS/DNA /  Capsid / Capsid /  Virion / Virion /  Parvovirus / Parvovirus /  Densovirus / Densovirus /  Invertebrate / Invertebrate /  Insect / Insect /  Pathogen / Pathogen /  ssDNA / ssDNA /  VIRUS / VIRUS-DNA complex VIRUS / VIRUS-DNA complex | |||||||||
| Function / homology |  DNA DNA Function and homology information Function and homology information | |||||||||
| Biological species |  Zophobas morio black wasting virus Zophobas morio black wasting virus | |||||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2 Å cryo EM / Resolution: 2 Å | |||||||||
 Authors Authors | Penzes, J.J. / Kaelber, J.T. | |||||||||
| Funding support |  United States, 2items United States, 2items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Sequencing-free discovery by cryo-EM of a pathogenic parvovirus causing mass mortality of farmed beetles Authors: Penzes, J.J. / Holm, M. / Firlar, E. / Kaelber, J.T. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8t9x.cif.gz 8t9x.cif.gz | 170 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8t9x.ent.gz pdb8t9x.ent.gz | 133.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8t9x.json.gz 8t9x.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/t9/8t9x https://data.pdbj.org/pub/pdb/validation_reports/t9/8t9x ftp://data.pdbj.org/pub/pdb/validation_reports/t9/8t9x ftp://data.pdbj.org/pub/pdb/validation_reports/t9/8t9x | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  41125MC  8t9cC 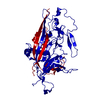 8t9eC  8ta7C  8tjeC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 | x 60
|
| 2 |
|
| 3 | x 5
|
| 4 | x 6
|
| 5 | 
|
| Symmetry | Point symmetry: (Schoenflies symbol : I (icosahedral : I (icosahedral )) )) |
- Components
Components
| #1: Protein | Mass: 48085.629 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Zophobas morio black wasting virus Zophobas morio black wasting virus |
|---|---|
| #2: DNA chain | Mass: 886.637 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Zophobas morio black wasting virus Zophobas morio black wasting virus |
| #3: DNA chain | Mass: 2733.827 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Zophobas morio black wasting virus Zophobas morio black wasting virus |
| #4: DNA chain | Mass: 1504.037 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Zophobas morio black wasting virus Zophobas morio black wasting virus |
| #5: Water | ChemComp-HOH /  Water Water |
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Zophobas morio densovirus / Type: VIRUS Details: Purified from T. molitor larvae, which were asymptomatic Entity ID: #1-#4 / Source: NATURAL |
|---|---|
| Molecular weight | Experimental value: NO |
| Source (natural) | Organism:  Zophobas morio densovirus / Strain: NJ2-molitor Zophobas morio densovirus / Strain: NJ2-molitor |
| Details of virus | Empty: NO / Enveloped: NO / Isolate: SPECIES / Type: VIRION |
| Natural host | Organism: Tenebrio molitor |
| Virus shell | Diameter: 28 nm / Triangulation number (T number): 1 |
| Buffer solution | pH: 7.4 |
| Buffer component | Conc.: 1 x / Name: Phosphate-buffered saline / Formula: PBS |
| Specimen | Conc.: 2 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YESDetails: Purified virus from homogenized T. molitor larval tissue |
| Specimen support | Grid material: GOLD / Grid mesh size: 300 divisions/in. / Grid type: UltrAuFoil R1.2/1.3 |
Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 95 % / Chamber temperature: 293 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TALOS ARCTICA |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 200 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 200 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1200 nm / Nominal defocus min: 500 nm / Cs Bright-field microscopy / Nominal defocus max: 1200 nm / Nominal defocus min: 500 nm / Cs : 2.7 mm : 2.7 mm |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 3 sec. / Electron dose: 32 e/Å2 / Detector mode: COUNTING / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Num. of grids imaged: 1 |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 156329 | ||||||||||||||||||||||||||||
| Symmetry | Point symmetry : I (icosahedral : I (icosahedral ) ) | ||||||||||||||||||||||||||||
3D reconstruction | Resolution: 2 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 15964 / Symmetry type: POINT | ||||||||||||||||||||||||||||
| Atomic model building | Protocol: AB INITIO MODEL / Space: REAL | ||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj

