[English] 日本語
 Yorodumi
Yorodumi- PDB-8r6y: Structure of the SFTSV L protein stalled in a transcription-speci... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8r6y | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the SFTSV L protein stalled in a transcription-specific early elongation state with bound capped RNA [TRANSCRIPTION-EARLY-ELONGATION] | ||||||||||||
 Components Components |
| ||||||||||||
 Keywords Keywords |  VIRAL PROTEIN / VIRAL PROTEIN /  SFTSV / SFTSV /  RNA-DEPENDENT RNA POLYMERASE / RNA-DEPENDENT RNA POLYMERASE /  VIRAL RNA VIRAL RNA | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell endoplasmic reticulum /  endoplasmic reticulum-Golgi intermediate compartment / host cell endoplasmic reticulum-Golgi intermediate compartment / host cell Golgi apparatus / endoplasmic reticulum-Golgi intermediate compartment / host cell endoplasmic reticulum-Golgi intermediate compartment / host cell Golgi apparatus /  RNA-directed RNA polymerase / viral RNA genome replication / RNA-directed RNA polymerase / viral RNA genome replication /  RNA-dependent RNA polymerase activity / DNA-templated transcription / RNA-dependent RNA polymerase activity / DNA-templated transcription /  metal ion binding metal ion bindingSimilarity search - Function | ||||||||||||
| Biological species |   SFTS virus AH12 SFTS virus AH12 | ||||||||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.4 Å cryo EM / Resolution: 3.4 Å | ||||||||||||
 Authors Authors | Williams, H.M. / Thorkelsson, S.R. / Vogel, D. / Busch, C. / Milewski, M. / Cusack, S. / Grunewald, K. / Quemin, E.R.J. / Rosenthal, M. | ||||||||||||
| Funding support |  Germany, 3items Germany, 3items
| ||||||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2024 Journal: Nucleic Acids Res / Year: 2024Title: Structural snapshots of phenuivirus cap-snatching and transcription. Authors: Harry M Williams / Sigurdur R Thorkelsson / Dominik Vogel / Carola Busch / Morlin Milewski / Stephen Cusack / Kay Grünewald / Emmanuelle R J Quemin / Maria Rosenthal /   Abstract: Severe fever with thrombocytopenia syndrome virus (SFTSV) is a human pathogen that is now endemic to several East Asian countries. The viral large (L) protein catalyzes viral transcription by ...Severe fever with thrombocytopenia syndrome virus (SFTSV) is a human pathogen that is now endemic to several East Asian countries. The viral large (L) protein catalyzes viral transcription by stealing host mRNA caps via a process known as cap-snatching. Here, we establish an in vitro cap-snatching assay and present three high-quality electron cryo-microscopy (cryo-EM) structures of the SFTSV L protein in biologically relevant, transcription-specific states. In a priming-state structure, we show capped RNA bound to the L protein cap-binding domain (CBD). The L protein conformation in this priming structure is significantly different from published replication-state structures, in particular the N- and C-terminal domains. The capped-RNA is positioned in a way that it can feed directly into the RNA-dependent RNA polymerase (RdRp) ready for elongation. We also captured the L protein in an early-elongation state following primer-incorporation demonstrating that this priming conformation is retained at least in the very early stages of primer extension. This structural data is complemented by in vitro biochemical and cell-based assays. Together, these insights further our mechanistic understanding of how SFTSV and other bunyaviruses incorporate stolen host mRNA fragments into their viral transcripts thereby allowing the virus to hijack host cell translation machinery. #1:  Journal: BiorXiv / Year: 2023 Journal: BiorXiv / Year: 2023Title: Structural snapshots of phenuivirus cap-snatching and transcription Authors: Williams, H.M. / Thorkelsson, S.R. / Vogel, D. / Busch, C. / Milewski, M. / Cusack, S. / Grunewald, K. / Quemin, E.R.J. / Rosenthal, M. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8r6y.cif.gz 8r6y.cif.gz | 432.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8r6y.ent.gz pdb8r6y.ent.gz | 338 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8r6y.json.gz 8r6y.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/r6/8r6y https://data.pdbj.org/pub/pdb/validation_reports/r6/8r6y ftp://data.pdbj.org/pub/pdb/validation_reports/r6/8r6y ftp://data.pdbj.org/pub/pdb/validation_reports/r6/8r6y | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  18969MC  8r6uC  8r6wC C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 235698.500 Da / Num. of mol.: 1 / Mutation: D112A Source method: isolated from a genetically manipulated source Source: (gene. exp.)   SFTS virus AH12 / Production host: SFTS virus AH12 / Production host:   Trichoplusia ni (cabbage looper) / References: UniProt: U3GU88 Trichoplusia ni (cabbage looper) / References: UniProt: U3GU88 |
|---|
-RNA chain , 4 types, 4 molecules PTGC
| #2: RNA chain | Mass: 6451.975 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: Synthesised by Biomers. / Source: (synth.)   SFTS virus AH12 SFTS virus AH12 |
|---|---|
| #3: RNA chain | Mass: 6480.872 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: Synthesised by Biomers. / Source: (synth.)   SFTS virus AH12 SFTS virus AH12 |
| #4: RNA chain | Mass: 918.636 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: Produced by the viral polymerase - not synthesised. Source: (synth.)   SFTS virus AH12 SFTS virus AH12 |
| #5: RNA chain | Mass: 5863.617 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: Synthesised by ChemGenes. / Source: (synth.)   SFTS virus AH12 SFTS virus AH12 |
-Non-polymers , 2 types, 2 molecules 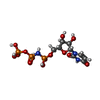


| #6: Chemical | ChemComp-2KH / |
|---|---|
| #7: Chemical | ChemComp-MG / |
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: SFTSV L protein in complex with 5' and 3' RNA, as well as RNA acting as a primer. Type: COMPLEX Details: SFTSV L protein expressed recombinantly in complex with several RNA species. Entity ID: #1-#4 / Source: RECOMBINANT |
|---|---|
| Molecular weight | Value: 0.24 MDa / Experimental value: YES |
| Source (natural) | Organism:   SFTS virus AH12 SFTS virus AH12 |
| Source (recombinant) | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Buffer solution | pH: 7 |
| Specimen | Conc.: 0.8 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES / Details: This sample was monodisperse. : YES / Details: This sample was monodisperse. |
| Specimen support | Film material: CARBON / Grid material: GOLD / Grid type: Quantifoil R2/1 |
Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE-PROPANE / Humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal magnification: 130000 X / Nominal defocus max: 2600 nm / Nominal defocus min: 800 nm / Cs Bright-field microscopy / Nominal magnification: 130000 X / Nominal defocus max: 2600 nm / Nominal defocus min: 800 nm / Cs : 2.7 mm : 2.7 mm |
| Specimen holder | Cryogen: NITROGEN |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Num. of real images: 27095 |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 2998852 | ||||||||||||||||||||||||||||||
3D reconstruction | Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 264083 / Symmetry type: POINT | ||||||||||||||||||||||||||||||
| Atomic model building | Protocol: RIGID BODY FIT / Target criteria: Cross-correlation coefficient | ||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj































