+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7yrg | ||||||
|---|---|---|---|---|---|---|---|
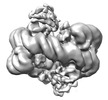
| Title | histone methyltransferase | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  GENE REGULATION / GENE REGULATION /  histone methyltransferase histone methyltransferase | ||||||
| Function / homology |  Function and homology information Function and homology informationhistone H4K20me methyltransferase activity / [histone H4]-N-methyl-L-lysine20 N-methyltransferase / histone H4K20 monomethyltransferase activity / [histone H4]-lysine20 N-methyltransferase / muscle organ development / heterochromatin organization / nucleosomal DNA binding / RNA polymerase II core promoter sequence-specific DNA binding /  heterochromatin / cellular response to estradiol stimulus ...histone H4K20me methyltransferase activity / [histone H4]-N-methyl-L-lysine20 N-methyltransferase / histone H4K20 monomethyltransferase activity / [histone H4]-lysine20 N-methyltransferase / muscle organ development / heterochromatin organization / nucleosomal DNA binding / RNA polymerase II core promoter sequence-specific DNA binding / heterochromatin / cellular response to estradiol stimulus ...histone H4K20me methyltransferase activity / [histone H4]-N-methyl-L-lysine20 N-methyltransferase / histone H4K20 monomethyltransferase activity / [histone H4]-lysine20 N-methyltransferase / muscle organ development / heterochromatin organization / nucleosomal DNA binding / RNA polymerase II core promoter sequence-specific DNA binding /  heterochromatin / cellular response to estradiol stimulus / heterochromatin / cellular response to estradiol stimulus /  euchromatin / chromatin DNA binding / structural constituent of chromatin / euchromatin / chromatin DNA binding / structural constituent of chromatin /  nucleosome / nucleosome /  nucleosome assembly / chromatin organization / RNA polymerase II cis-regulatory region sequence-specific DNA binding / protein heterodimerization activity / Amyloid fiber formation / positive regulation of transcription by RNA polymerase II / nucleosome assembly / chromatin organization / RNA polymerase II cis-regulatory region sequence-specific DNA binding / protein heterodimerization activity / Amyloid fiber formation / positive regulation of transcription by RNA polymerase II /  DNA binding / extracellular exosome / DNA binding / extracellular exosome /  nucleoplasm / nucleoplasm /  nucleus nucleusSimilarity search - Function | ||||||
| Biological species |  Xenopus laevis (African clawed frog) Xenopus laevis (African clawed frog)  Homo sapiens (human) Homo sapiens (human) | ||||||
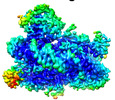
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.2 Å cryo EM / Resolution: 4.2 Å | ||||||
 Authors Authors | Li, H. / Wang, W.Y. | ||||||
| Funding support |  China, 1items China, 1items
| ||||||
 Citation Citation |  Journal: Mol Cell / Year: 2023 Journal: Mol Cell / Year: 2023Title: Structural insight into H4K20 methylation on H2A.Z-nucleosome by SUV420H1. Authors: Li Huang / Youwang Wang / Haizhen Long / Haoqiang Zhu / Zengqi Wen / Liwei Zhang / Wenhao Zhang / Zhenqian Guo / Longge Wang / Fangyi Tang / Jie Hu / Keyan Bao / Ping Zhu / Guohong Li / Zheng Zhou /  Abstract: DNA replication ensures the accurate transmission of genetic information during the cell cycle. Histone variant H2A.Z is crucial for early replication origins licensing and activation in which ...DNA replication ensures the accurate transmission of genetic information during the cell cycle. Histone variant H2A.Z is crucial for early replication origins licensing and activation in which SUV420H1 preferentially recognizes H2A.Z-nucleosome and deposits H4 lysine 20 dimethylation (H4K20me2) on replication origins. Here, we report the cryo-EM structures of SUV420H1 bound to H2A.Z-nucleosome or H2A-nucleosome and demonstrate that SUV420H1 directly interacts with H4 N-terminal tail, the DNA, and the acidic patch in the nucleosome. The H4 (1-24) forms a lasso-shaped structure that stabilizes the SUV420H1-nucleosome complex and precisely projects the H4K20 residue into the SUV420H1 catalytic center. In vitro and in vivo analyses reveal a crucial role of the SUV420H1 KR loop (residues 214-223), which lies close to the H2A.Z-specific residues D97/S98, in H2A.Z-nucleosome preferential recognition. Together, our findings elucidate how SUV420H1 recognizes nucleosomes to ensure site-specific H4K20me2 modification and provide insights into how SUV420H1 preferentially recognizes H2A.Z nucleosome. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7yrg.cif.gz 7yrg.cif.gz | 378.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7yrg.ent.gz pdb7yrg.ent.gz | 291 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7yrg.json.gz 7yrg.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/yr/7yrg https://data.pdbj.org/pub/pdb/validation_reports/yr/7yrg ftp://data.pdbj.org/pub/pdb/validation_reports/yr/7yrg ftp://data.pdbj.org/pub/pdb/validation_reports/yr/7yrg | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  34055MC  7yrdC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 5 types, 10 molecules AEBFCGDHKL
| #1: Protein | Mass: 12046.091 Da / Num. of mol.: 2 / Mutation: G2E,G70A Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Xenopus laevis (African clawed frog) Xenopus laevis (African clawed frog)Production host:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria)References: UniProt: P84233 #2: Protein |  Mass: 11265.247 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Xenopus laevis (African clawed frog) Xenopus laevis (African clawed frog)Production host:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria)References: UniProt: P62799 #4: Protein |  / H2A/z / H2A/zMass: 12091.093 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: H2AZ1, H2AFZ, H2AZ Homo sapiens (human) / Gene: H2AZ1, H2AFZ, H2AZProduction host:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria)References: UniProt: P0C0S5 #5: Protein | Mass: 10607.212 Da / Num. of mol.: 2 / Mutation: S33T Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Xenopus laevis (African clawed frog) Xenopus laevis (African clawed frog)Production host:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria)References: UniProt: P02281 #6: Protein | Mass: 32214.787 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: KMT5B Homo sapiens (human) / Gene: KMT5BProduction host:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria)References: UniProt: B7WNX0, [histone H4]-lysine20 N-methyltransferase, [histone H4]-N-methyl-L-lysine20 N-methyltransferase |
|---|
-DNA chain , 1 types, 2 molecules IJ
| #3: DNA chain | Mass: 45054.844 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) Homo sapiens (human)Production host:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) |
|---|
-Non-polymers , 2 types, 4 molecules 


| #7: Chemical | | #8: Chemical |  S-Adenosyl methionine S-Adenosyl methionine |
|---|
-Details
| Has ligand of interest | N |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source (natural) |
| ||||||||||||||||||||||||
| Source (recombinant) | Organism:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) | ||||||||||||||||||||||||
| Buffer solution | pH: 7 | ||||||||||||||||||||||||
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES | ||||||||||||||||||||||||
Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TALOS ARCTICA |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 200 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 200 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2300 nm / Nominal defocus min: 1800 nm Bright-field microscopy / Nominal defocus max: 2300 nm / Nominal defocus min: 1800 nm |
| Image recording | Electron dose: 60 e/Å2 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
- Processing
Processing
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
3D reconstruction | Resolution: 4.2 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 77000 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller




 PDBj
PDBj








































