+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | RNA origami 3-helix tile Traptamer | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords |  Origami / Origami /  aptamer / aptamer /  switch / switch /  sensor / toe-hold / robot / broccoli / sensor / toe-hold / robot / broccoli /  RNA RNA | |||||||||||||||||||||
| Biological species | synthetic construct (others) | |||||||||||||||||||||
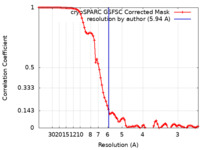
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 5.94 Å cryo EM / Resolution: 5.94 Å | |||||||||||||||||||||
 Authors Authors | McRae EKS / Vallina NS / Andersen ES | |||||||||||||||||||||
| Funding support | European Union,  Denmark, Denmark,  Canada, 6 items Canada, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2024 Journal: Sci Adv / Year: 2024Title: An RNA origami robot that traps and releases a fluorescent aptamer. Authors: Néstor Sampedro Vallina / Ewan K S McRae / Cody Geary / Ebbe S Andersen /   Abstract: RNA nanotechnology aims to use RNA as a programmable material to create self-assembling nanodevices for application in medicine and synthetic biology. The main challenge is to develop advanced RNA ...RNA nanotechnology aims to use RNA as a programmable material to create self-assembling nanodevices for application in medicine and synthetic biology. The main challenge is to develop advanced RNA robotic devices that both sense, compute, and actuate to obtain enhanced control over molecular processes. Here, we use the RNA origami method to prototype an RNA robotic device, named the "Traptamer," that mechanically traps the fluorescent aptamer, iSpinach. The Traptamer is shown to sense two RNA key strands, acts as a Boolean AND gate, and reversibly controls the fluorescence of the iSpinach aptamer. Cryo-electron microscopy of the closed Traptamer structure at 5.45-angstrom resolution reveals the mechanical mode of distortion of the iSpinach motif. Our study suggests a general approach to distorting RNA motifs and a path forward to build sophisticated RNA machines that through sensing, computing, and actuation modules can be used to precisely control RNA functionalities in cellular systems. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41656.map.gz emd_41656.map.gz | 31.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41656-v30.xml emd-41656-v30.xml emd-41656.xml emd-41656.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41656_fsc.xml emd_41656_fsc.xml | 9.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_41656.png emd_41656.png | 70 KB | ||
| Masks |  emd_41656_msk_1.map emd_41656_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-41656.cif.gz emd-41656.cif.gz | 5.5 KB | ||
| Others |  emd_41656_additional_1.map.gz emd_41656_additional_1.map.gz emd_41656_half_map_1.map.gz emd_41656_half_map_1.map.gz emd_41656_half_map_2.map.gz emd_41656_half_map_2.map.gz | 57.5 MB 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41656 http://ftp.pdbj.org/pub/emdb/structures/EMD-41656 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41656 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41656 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_41656.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41656.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.294 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_41656_msk_1.map emd_41656_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_41656_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_41656_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_41656_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 14-14 Traptamer
| Entire | Name: 14-14 Traptamer |
|---|---|
| Components |
|
-Supramolecule #1: 14-14 Traptamer
| Supramolecule | Name: 14-14 Traptamer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: An RNA origami 3 helix tile with an inactive broccoli aptamer in the central helix. |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: RNA (363-MER)
| Macromolecule | Name: RNA (363-MER) / type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 116.977742 KDa |
| Sequence | String: GGAAUCUCGC CCGAUGUUCG CAUCGGGAUU UGCAGGUCCA UGGAUUACAC CAUGCAACGC AGACCUGUAG AUGCCACGCU AGCCGUGGU GAGGGUCGGG UCCAGAUGUC AUUCGACUUU AACGCGCCUA AGCGUUGAAG GCGUGUUAGA GCAGAUAGUU C GCUAUCUG ...String: GGAAUCUCGC CCGAUGUUCG CAUCGGGAUU UGCAGGUCCA UGGAUUACAC CAUGCAACGC AGACCUGUAG AUGCCACGCU AGCCGUGGU GAGGGUCGGG UCCAGAUGUC AUUCGACUUU AACGCGCCUA AGCGUUGAAG GCGUGUUAGA GCAGAUAGUU C GCUAUCUG GGGAGCCUGU UCGCAGGCUC AGGAGCCUUC GGGCUCCUAG CGCUAUUACC CCGGACACCA CCGGGCAGAC AA GUAAUGG UGCUCCUCGA AUGACUUCUG UUGAGUAGAG UGUGGGCUCC GCGGCUAGUG UGCACCUUAG CGGUGAAUGU CUG ACACCG UUAAGGUGGU UACUCUUCGG AGUAACGCCG AGAUUCC |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: AIR / Details: 15mA | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 294 K / Instrument: LEICA EM GP / Details: Protochips Au-Flat 1.2/1.3 grids.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Average electron dose: 60.0 e/Å2 / Details: Pixel size of 0.645 A/px |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X