+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of IRL1620-bound ETBR-Gi complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | IRL1620 / ETBR / Gi /  MEMBRANE PROTEIN MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationenteric smooth muscle cell differentiation / response to endothelin /  endothelin receptor activity / negative regulation of neuron maturation / aldosterone metabolic process / chordate pharynx development / vein smooth muscle contraction / regulation of fever generation / heparin metabolic process / positive regulation of penile erection ...enteric smooth muscle cell differentiation / response to endothelin / endothelin receptor activity / negative regulation of neuron maturation / aldosterone metabolic process / chordate pharynx development / vein smooth muscle contraction / regulation of fever generation / heparin metabolic process / positive regulation of penile erection ...enteric smooth muscle cell differentiation / response to endothelin /  endothelin receptor activity / negative regulation of neuron maturation / aldosterone metabolic process / chordate pharynx development / vein smooth muscle contraction / regulation of fever generation / heparin metabolic process / positive regulation of penile erection / neuroblast migration / posterior midgut development / epithelial fluid transport / podocyte differentiation / protein transmembrane transport / endothelin receptor signaling pathway / developmental pigmentation / renal sodium ion absorption / response to sodium phosphate / endothelin receptor activity / negative regulation of neuron maturation / aldosterone metabolic process / chordate pharynx development / vein smooth muscle contraction / regulation of fever generation / heparin metabolic process / positive regulation of penile erection / neuroblast migration / posterior midgut development / epithelial fluid transport / podocyte differentiation / protein transmembrane transport / endothelin receptor signaling pathway / developmental pigmentation / renal sodium ion absorption / response to sodium phosphate /  enteric nervous system development / renal sodium excretion / renin secretion into blood stream / renal albumin absorption / melanocyte differentiation / regulation of pH / positive regulation of urine volume / negative regulation of adenylate cyclase activity / enteric nervous system development / renal sodium excretion / renin secretion into blood stream / renal albumin absorption / melanocyte differentiation / regulation of pH / positive regulation of urine volume / negative regulation of adenylate cyclase activity /  peripheral nervous system development / peripheral nervous system development /  vasoconstriction / regulation of epithelial cell proliferation / type 1 angiotensin receptor binding / establishment of endothelial barrier / vasoconstriction / regulation of epithelial cell proliferation / type 1 angiotensin receptor binding / establishment of endothelial barrier /  neural crest cell migration / negative regulation of protein metabolic process / response to pain / macrophage chemotaxis / cGMP-mediated signaling / neural crest cell migration / negative regulation of protein metabolic process / response to pain / macrophage chemotaxis / cGMP-mediated signaling /  peptide hormone binding / Adenylate cyclase inhibitory pathway / canonical Wnt signaling pathway / positive regulation of protein localization to cell cortex / regulation of cAMP-mediated signaling / D2 dopamine receptor binding / G protein-coupled serotonin receptor binding / regulation of mitotic spindle organization / cellular response to forskolin / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / peptide hormone binding / Adenylate cyclase inhibitory pathway / canonical Wnt signaling pathway / positive regulation of protein localization to cell cortex / regulation of cAMP-mediated signaling / D2 dopamine receptor binding / G protein-coupled serotonin receptor binding / regulation of mitotic spindle organization / cellular response to forskolin / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway /  regulation of heart rate / Peptide ligand-binding receptors / Regulation of insulin secretion / G protein-coupled receptor binding / calcium-mediated signaling / calcium ion transmembrane transport / Olfactory Signaling Pathway / G-protein beta/gamma-subunit complex binding / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / response to organic cyclic compound / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / Sensory perception of sweet, bitter, and umami (glutamate) taste / photoreceptor disc membrane / response to peptide hormone / Adrenaline,noradrenaline inhibits insulin secretion / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / cellular response to catecholamine stimulus / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / regulation of heart rate / Peptide ligand-binding receptors / Regulation of insulin secretion / G protein-coupled receptor binding / calcium-mediated signaling / calcium ion transmembrane transport / Olfactory Signaling Pathway / G-protein beta/gamma-subunit complex binding / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / response to organic cyclic compound / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / Sensory perception of sweet, bitter, and umami (glutamate) taste / photoreceptor disc membrane / response to peptide hormone / Adrenaline,noradrenaline inhibits insulin secretion / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / cellular response to catecholamine stimulus / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding /  vasodilation / sensory perception of taste / GPER1 signaling / G-protein beta-subunit binding / GDP binding / Inactivation, recovery and regulation of the phototransduction cascade / vasodilation / sensory perception of taste / GPER1 signaling / G-protein beta-subunit binding / GDP binding / Inactivation, recovery and regulation of the phototransduction cascade /  heterotrimeric G-protein complex / G alpha (12/13) signalling events / heterotrimeric G-protein complex / G alpha (12/13) signalling events /  extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye /  GTPase binding / Ca2+ pathway GTPase binding / Ca2+ pathwaySimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /   Mus musculus (house mouse) Mus musculus (house mouse) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.99 Å cryo EM / Resolution: 2.99 Å | |||||||||
 Authors Authors | Yuan Q / Ji Y / Jiang Y / Duan J / Xu HE | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural basis of peptide recognition and activation of endothelin receptors. Authors: Yujie Ji / Jia Duan / Qingning Yuan / Xinheng He / Gong Yang / Shengnan Zhu / Kai Wu / Wen Hu / Tianyu Gao / Xi Cheng / Hualiang Jiang / H Eric Xu / Yi Jiang /  Abstract: Endothelin system comprises three endogenous 21-amino-acid peptide ligands endothelin-1, -2, and -3 (ET-1/2/3), and two G protein-coupled receptor (GPCR) subtypes-endothelin receptor A (ETR) and B ...Endothelin system comprises three endogenous 21-amino-acid peptide ligands endothelin-1, -2, and -3 (ET-1/2/3), and two G protein-coupled receptor (GPCR) subtypes-endothelin receptor A (ETR) and B (ETR). Since ET-1, the first endothelin, was identified in 1988 as one of the most potent endothelial cell-derived vasoconstrictor peptides with long-lasting actions, the endothelin system has attracted extensive attention due to its critical role in vasoregulation and close relevance in cardiovascular-related diseases. Here we present three cryo-electron microscopy structures of ETR and ETR bound to ET-1 and ETR bound to the selective peptide IRL1620. These structures reveal a highly conserved recognition mode of ET-1 and characterize the ligand selectivity by ETRs. They also present several conformation features of the active ETRs, thus revealing a specific activation mechanism. Together, these findings deepen our understanding of endothelin system regulation and offer an opportunity to design selective drugs targeting specific ETR subtypes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34619.map.gz emd_34619.map.gz | 57.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34619-v30.xml emd-34619-v30.xml emd-34619.xml emd-34619.xml | 20 KB 20 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34619.png emd_34619.png | 36.9 KB | ||
| Filedesc metadata |  emd-34619.cif.gz emd-34619.cif.gz | 6.7 KB | ||
| Others |  emd_34619_half_map_1.map.gz emd_34619_half_map_1.map.gz emd_34619_half_map_2.map.gz emd_34619_half_map_2.map.gz | 49.5 MB 49.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34619 http://ftp.pdbj.org/pub/emdb/structures/EMD-34619 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34619 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34619 | HTTPS FTP |
-Related structure data
| Related structure data |  8hbdMC  8hcqC  8hcxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34619.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34619.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34619_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
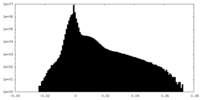
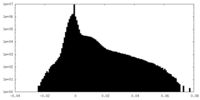
| Projections & Slices |
| ||||||||||||
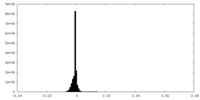
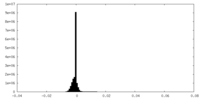
| Density Histograms |
-Half map: #1
| File | emd_34619_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : IRL1620-bound ETBR-Gi complex
| Entire | Name: IRL1620-bound ETBR-Gi complex |
|---|---|
| Components |
|
-Supramolecule #1: IRL1620-bound ETBR-Gi complex
| Supramolecule | Name: IRL1620-bound ETBR-Gi complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.445059 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGAQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHESM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCS TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.055867 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI ...String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI VTSSGDTTCA LWDIETGQQT TTFTGHTGDV MSLSLAPDTR LFVSGACDAS AKLWDVREGM CRQTFTGHES DI NAICFFP NGNAFATGSD DATCRLFDLR ADQELMTYSH DNIICGITSV SFSKSGRLLL AGYDDFNCNV WDALKADRAG VLA GHDNRV SCLGVTDDGM AVATGSWDSF LKIWNGSSGG GGSGGGGSSG VSGWRLFKKI S UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 30.363043 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MLLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFAVQ LVESGGGLVQ PGGSRKLSCS ASGFAFSSFG MHWVRQAPEK GLEWVAYIS SGSGTIYYAD TVKGRFTISR DDPKNTLFLQ MTSLRSEDTA MYYCVRSIYY YGSSPFDFWG QGTTLTVSAG G GGSGGGGS ...String: MLLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFAVQ LVESGGGLVQ PGGSRKLSCS ASGFAFSSFG MHWVRQAPEK GLEWVAYIS SGSGTIYYAD TVKGRFTISR DDPKNTLFLQ MTSLRSEDTA MYYCVRSIYY YGSSPFDFWG QGTTLTVSAG G GGSGGGGS GGGGSADIVM TQATSSVPVT PGESVSISCR SSKSLLHSNG NTYLYWFLQR PGQSPQLLIY RMSNLASGVP DR FSGSGSG TAFTLTISRL EAEDVGVYYC MQHLEYPLTF GAGTKLEL |
-Macromolecule #5: IRL1620
| Macromolecule | Name: IRL1620 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.83697 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: DDEEAVYFAH LDIIW |
-Macromolecule #6: Endothelin receptor type B,Endothelin receptor type B,Oplophorus-...
| Macromolecule | Name: Endothelin receptor type B,Endothelin receptor type B,Oplophorus-luciferin 2-monooxygenase catalytic subunit chimera type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO / EC number:  Oplophorus-luciferin 2-monooxygenase Oplophorus-luciferin 2-monooxygenase |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 67.88232 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MDSKGSSQKG SRLLLLLVVS NLLLCQGVVS DYKDDDDVDH HHHHHHHEER GFPPDRATPL LQTAEIMTPP TKTLWPKGSN ASLARSLAP AEVPKGDRTA GSPPRTISPP PCQGPIEIKE TFKYINTVVS CLVFVLGIIG NSTLLRIIYK NKCMRNGPNI L IASLALGD ...String: MDSKGSSQKG SRLLLLLVVS NLLLCQGVVS DYKDDDDVDH HHHHHHHEER GFPPDRATPL LQTAEIMTPP TKTLWPKGSN ASLARSLAP AEVPKGDRTA GSPPRTISPP PCQGPIEIKE TFKYINTVVS CLVFVLGIIG NSTLLRIIYK NKCMRNGPNI L IASLALGD LLHIVIDIPI NVYKLLAEDW PFGAEMCKLV PFIQKASVGI TVLSLCALSI DRYRAVASWS RIKGIGVPKW TA VEIVLIW VVSVVLAVPE AIGFDIITMD YKGSYLRICL LHPVQKTAFM QFYKTAKDWW LFSFYFCLPL AITAFFYTLM TCE MLRKKS GMQIALNDHL KQRREVAKTV FCLVLVFALC WLPLHLSRIL KLTLYNQNDP NRCELLSFLL VLDYIGINMA SLNS CINPI ALYLVSKRFK NCFKSCLCCW CQSFEEKQSL EEKQSCLKFK VFTLEDFVGD WEQTAAYNLD QVLEQGGVSS LLQNL AVSV TPIQRIVRSG ENALKIDIHV IIPYEGLSAD QMAQIEEVFK VVYPVDDHHF KVILPYGTLV IDGVTPNMLN YFGRPY EGI AVFDGKKITV TGTLWNGNKI IDERLITPDG SMLFRVTINS UniProtKB: Endothelin receptor type B |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.04 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 18.0 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Nominal defocus max: 18.0 µm / Nominal defocus min: 0.8 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.99 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 191493 |
 Movie
Movie Controller
Controller




































 Z
Z Y
Y X
X